 |
 |
- Search
| Psychiatry Investig > Volume 15(3); 2018 > Article |
|
Abstract
Objective
Autism spectrum disorders (ASD) have a complex pathophysiology including genetic, inflammatory and neurodevelopmental components. We aim to investigate the relationship between ASD and gene polymorphisms of stromal cell-derived factor-1 (SDF-1) and its receptor CXC chemokine receptor-4 (CXCR4), which may affect inflammatory and neurodevelopmental processes.
Methods
101 children diagnosed with ASD aged 2-18 and their biological parents were included in the study. All participants were assessed using an information form and the Children were assessed using Childhood Autism Rating Scale (CARS). SDF-1 G801→A and CXCR4 C13→T polymorphisms were detected by genetic techniques. The results were evaluated using the transmission disequilibrium test (TDT) and haplotype relative risk (HRR).
Results
Following TDT evaluation for CXCR4, the assumption of equality was not rejected (χ2=1.385, p=0.239). HRR for the C allele was 1.037 [HRR (95%CI)=0.937 (0.450-2.387), χ2=0.007, p=0.933] and HRR for the T allele was 0.965 [HRR (95%CI)=0.965 (0.419- 2.221), χ2=1.219, p=0.270], but the findings were statistically insignificant. Based on TDT evaluation for SDF1, the assumption of equality cannot be rejected (χ2=0, p=0.999). HRR for the A allele was 0.701 [HRR (95%CI)=0.701 (0.372-1.319), χ2=1.219, p=0.270] and HRR for the G allele was 1.427 [HRR (95%CI)=1.427 (0.758-2.686), χ2=1.219, p=0.270], but the findings were statistically insignificant.
Conclusion
The genetic screening of blood samples from mother, father and child trios could not show a significant association between SDF1/CXCR4 genes and ASD on the basis of TDT and HRR tests. More extensive genetic studies are now needed to investigate the relationship between SDF1/CXCR4 gene polymorphisms and ASD.
Autism spectrum disorder (ASD) is a neurodevelopmental condition characterized by impaired verbal and nonverbal communication skills, restricted social interaction, and repetitive or stereotyped behavioral characteristics. Studies are increasingly focusing on cellular changes such as modulation of the immune response, particularly caused by genetic-environmental interaction [1]. There is a consensus that the deviations in brain development in ASD are due to the with multiple mechanisms including immune dysfunction [2]. Immune system abnormalities such as irregularities in cytokine signaling are observed in ASD. Inflammation in brain tissue has been reported in post-mortem brain samples from individuals with ASD [3].
Chemokines regulate cell migration through receptors, as well as immunological responses, inflammation and infection. They also play an important role in the immune system. SDF1, a chemokine also known as CXCL12, has functions including T and B cell functions, and CD34+stem cell migration. The receptor of SDF1 is CXCR4 [4]. Chemokines provide binding and signal transduction to target cells via G protein receptors [5]. They also play an important role in the central nervous system and affect brain functions in harmony with the neurotransmitter and neuropeptide system [6]. SDF1/CXCR4 interaction is required for normal integration of the cerebral cortex [7]. SDF-1/CXCR4 has been shown in many areas within the central nervous system, such as astrocytes, neurons and microglia [8]. Death in the perinatal period was reported in mice lacking SDF1 in one study [9]. Another study investigating chemokine profile and ASD associations reported greater impairment of non-verbal cognitive ability and language comprehension skills in children with neonatal IL-4 elevation compared to a control group [10]. Elevated monocyte chemotactic protein-1 (MCP-1) and decreased Regulated upon Activation Normal T-Cell Expressed and Secreted (RANTES) levels were determined in the ASD cases compared to the control group [11]. In another study in which CXCR4 was experimentally excluded, mice exhibited stereotyped behaviors similar to ASD [12].
Genetic studies have revealed that ASD does not originate from a single cause. Different genetic disorders are known to cause the characteristic ASD phenotype through similar mechanisms [13]. IL1RAPL1 gene mutation encoding a protein affecting the interleukin-1 receptor that regulates cytokine activity has been associated with ASD [14]. A decreased 22q11.2 gene level disrupts the migration and positioning of neurons in the brain via CXCR4 [15]. Recently several study findings have pointed out the effects of single nucleotide polymorphisms (SNPs) in the etiology of several diseases. A SNP, CXCL12-G801A polymorphism, which increases SDF1 expression at position 801 (G to A) in the 3’-untranslated region (3’UTR) of SDF1, has been described [16]. CXCR4 is located on chromosome 2q2. SNP rs2228014 (C/T) was found at codon 138 [17]. This reveals that chemokines and chemokine-related genetic disorders may be involved in ASD. The aim of this study was to investigate the role of SDF-1 and CXCR4 gene polymorphism in the development of ASD.
The study group was selected from patients diagnosed with ASD at the Istanbul University Medical Faculty Department of Child and Adolescent Psychiatry, Turkey. Children diagnosed with ASD and aged 2-18 were included in the study together with their biological parents to ensure that participation was voluntary. Informed consent was obtained from all participants. One hundred one children and their biological parents were enrolled. All participants were evaluated using an information form. Patients also underwent psychiatric evaluation using DSM-IV and CARS. Blood samples from patients and their parents were classified in our clinic and sent to the Department of Molecular Medicine of the Istanbul University Institute for Experimental Medicine, where DNA was obtained within one working day at the latest.
The registration number of the study is 2012/1502-1229 and the approval of the Ethics Committee was taken on 12.10.2012.
Genomic DNA was obtained from patients and parents using standard salting-out techniques. Following isolation of DNA samples, the polymerase chain reaction (PCR) was performed with suitable primers and conditions (Table 1). PCR products were digested with an appropriate enzyme at 37℃ by restriction fragment length polymorphism (RFLP) analysis. The Bccl enzyme was used in the digestion of CXCR4 C-138T under conditions of 37℃ and a 2-hour incubation period. PvuII was used for SDF1 3’A under conditions of 37℃ and an overnight incubation period. The digested products were analyzed on 3% agarose gel stained with ethidium bromide. 236 bp, 133 and 103 bp bands were evaluated for CXCR4 C-138T (Figure 1). 302 bp, 201 and 101 bp were also evaluated for SDF1 3’A (Figure 2).
SNPs were obtained from genetic material from the study participants. All data were analyzed on the Number Cruncher Statistical System NCSS 2007 (NCSS LLC, Kaysville, UT, USA). Allelic and genotypic distributions of trios were tested using transmission disequilibrium test (TDT) and haplotype relative risk (HRR).
The study consisted of a total of 303 participants, including 101 children with OSB, 16 female and 85 male, and their biological parents. The children’s ages ranged from 3 to 18 years, with an average of 9.86. Kinship between parents was present in 19 cases and absent in 82. Comorbid psychiatric pathology was present in 67 (66.3%) children, while no comorbidities were identified in 34 (33.7%). When the children in the study were evaluated in terms of IQ scores, normal IQ values were determined in 7 (6.9%), borderline mental capacity in 8 (7.9%), mild mental retardation in 30 (29.7%), moderate mental retardation in 34 (33.7%), severe mental retardation in 20 (19.8%) and profound mental retardation in 2 (2.0%) patients. Constructed CARS values were calculated as a minimum of 30 and a maximum of 54.5 points, the average CARS score being 38.995.
CXCR4 evaluations were performed with 88 cases and the parents of these. TDT evaluation was performed only with data obtained from heterozygous parents (n=23). In 17 cases, the C allele was both transmitted and non-transmitted. In 16 cases, C transmitted, while T was non-transmitted. In 10 cases, T was transmitted and C was non-transmitted, while in 3 cases, T was both transmitted and non-transmitted. The TDT test was used to determine whether the proportions of C transmitted T nontransmitted cases and T transmitted C non-transmitted cases were equal. Based on that evaluation, the assumption of equality was not rejected (χ2=1.385, p=0.239) (Table 2).
According to the calculations performed with 88 cases, the HRR value for the C allele was determined at 1.037, but the analysis was statistically insignificant [HRR (95% CI)=1.037 (0.450-2.387), χ2=0.007, p=0.933]. The HRR value for the T-allele was 0.965, but the analysis was also statistically insignificant [HRR (95% CI)=0.965 (0.419-2.221), χ2=1.219, p=0.270].
SDF evaluations were performed with 101 cases and the parents thereof. TDT evaluation was performed only with data obtained from heterozygous parents (n=56). In 3 cases, the A allele was both transmitted and non-transmitted. In 34 cases, A was transmitted and G was non-transmitted. In 34 cases, G was transmitted and A was non-transmitted, while in 41 cases, G was both transmitted and non-transmitted. The TDT test was used to determine whether the proportions of A transmitted G non-transmitted cases and G transmitted A non-transmitted cases were equal. Based on that evaluation, the assumption of equality was not rejected (χ2=0, p=0.999) (Table 3).
According to calculation made with all 101 cases, the HRR value for the A allele was 0.701, but the analysis was statistically insignificant [HRR (95% CI)=0.701 (0.372-1.319), χ2=1.219, p=0.270]. The HRR value for the G allele was 1.427, but the analysis was also statistically insignificant [HRR (95% CI)=1.427 (0.758-2.686), χ2=1.219, p=0.270].
Previous studies have evaluated the role of chemokines and chemokine receptors in psychiatric diseases, but none have involved ASD-associated SDF-1/CXCR4 gene polymorphism. To the best of our knowledge, this is the first study of SDF-1 G801→A (also known as SDF-1 3’A) and CXCR4 C138→T polymorphism in ASD.
Numerous studies have investigated the presence and function of chemokines in the central nervous system. SDF1 and its receptor CXCR4 also function outside the immune system, and defects are seen in various organs, including cerebellar development, when these are deficient [18]. Chemokines and their receptors are known to contribute to homeostasis in the brain [19]. Experiments with rats have shown the presence of genes encoding CXCR4 in cells and tissues of the central nervous system [20]. Chemokines and receptors have been shown to play an important role in nervous system damage [21]. Chemokines are also involved in cell migration during embryological development. Animals in which CXCR4 was not active died shortly after birth and exhibited hematopoietic and neurological defects. SDF-1/CXCR-4 interaction has been shown to be involved in regulating neuronal cell migration [22]. Neural stem/progenitor cells excrete chemokine receptors. Neuronal stem cell migration has been shown to be impaired by blocking chemokines and ligands that are detected in the brain [23]. CXCR4 positive neural progenitor cells have been associated with neurodevelopment and neuroplasticity in bipolar disorder [24]. These studies provide strong evidence for the relationship between chemokines and the central nervous system, and it appears that chemokines play an important role in this.
Studies of the immune system in ASD indicate that many immunological markers may be associated with autism. Immune system anomalies such as increased autoimmunity or decreased immunity have been reported in children with ASD. The hypothesis of immune system disorders in ASD is based on the known role of the immune system in early neurodevelopment and the effect of these impairments on behavior patterns [25]. One study investigating immunological determinants in high-functioning autistics reported significantly higher IL-1beta, IL-1RA, IL-5, IL-8, IL-12 (p70), IL-13, IL-17, and GROalpha plasma concentrations compared to a control group [26]. Altered cell-mediated immunity may also be expected to play a role in the early neonatal period in ASD. It has been suggested that high levels of MCP-1 in the amniotic fluid constitute a high risk for ASD and that MCP-1 may be involved in the pathophysiology of ASD. Reduced levels of RANTES in newborn blood samples have been found to be associated with the development of ASD in later life [27]. In a study of children with ASD by Ashwood et al. [28], increased chemokine production was found to be associated with high abnormal behavior scores and impaired developmental and adaptive functions. Studies that we have reviewed report that immunological changes associated with chemokines contribute to the risk of developing ASD and affect behavior patterns.
Genetic studies have increased in recent years, and many genetic variants have been associated with diseases. There have been many studies of SDF1 and CXCR4 gene polymorphisms. SDF1-3’A gene polymorphism has been linked to the progression of prostate cancer and bone metastasis [29]. SDF1 and CXCR4 polymorphisms have also been shown to have a significant association with susceptibility and prognosis for some types of lung cancer [30]. SDF1 gene polymorphism has been associated with age at onset of type 1 diabetes [31]. While SDF1 and CXCR4 gene polymorphisms have not been studied in psychiatric disorders, many other genetic variation studies have reported that many single nucleotide polymorphisms are related to psychiatric disorders. Many SNP studies have been performed in the context of ASD. Some studies have reported that SNPs are directly related to the etiology of ASD, while others have associated them with findings and symptom severity seen in ASD [32-34]. In a study investigating variations in copy number and linkage analysis in autism, chemokine pathways and developmental pathways were identified as two similar molecular themes in two independent genome-scale measurements [35], Studies focusing on candidate genes have shown that several genes associated with neuronal synaptic function play a role in the development of ASD. Some single-nucleotide polymorphisms (SNP) associated with ASD have been identified in linkage studies [36]. In one study, SNPs, which may be associated with language delay in ASD, were identified on chromosome 11 and assessed using TDT [37]. Genetic mechanisms may play a role in the regulation of the immune system, and some studies have suggested that immunomodulatory genes in chromosome 6 may be linked to autism [38]. One study conducted with TDT showed that HLA-DR4 and DR13 are associated with ASD [39].
The studies we investigated evaluated the relationship between ASD and SNPs using several different statistical methods. Genetic studies of this kind may have some limitations. Association samples are also prone to type 2 errors. In general, it has been suggested that negative results may occur from type 2 errors due to fewer samples being taken as needed [40]. Type 2 statistical errors reject the existing relationship between groups. Although there is a difference between the groups we investigate, we can still find that “there is no difference.” More extensive studies should now be performed in order to confirm this. In this study, we observed no significant correlation between these two genetic polymorphisms and ASD. However, it would be useful to repeat this study with a larger sample size and using more genetic markers. More extensive studies are now needed to test the relationship between ASD and the SDF-1 and CXCR4 genes.
ACKNOWLEDGEMENTS
We are grateful to the Scientific Research Projects Coordination Unit for providing financial support. This research was supported by the Scientific Research Projects Coordination Unit of Istanbul University (Project number: 29031).
Figure 1.
CXCR4 C-138T gene variants. C: Cytosine, T: Thymine, CXCR4: receptor CXC chemokine receptor-4, bp: Base pairs.
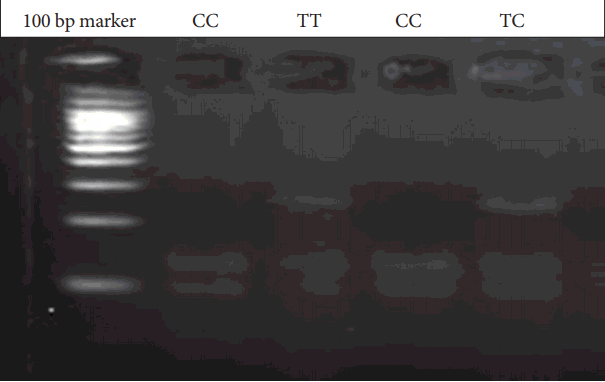
Figure 2.
SDF1 3’A gene variants. A: Adenine, G:Guanine, SDF: stromal cell-derived factor, bp: Base pairs.
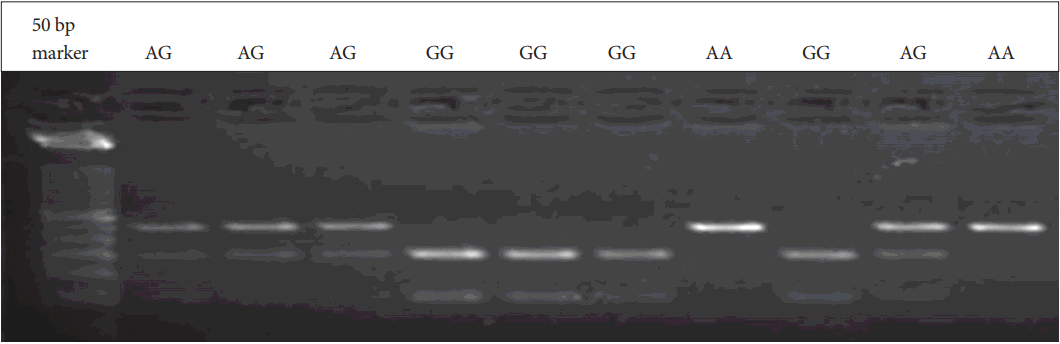
Table 1.
Gene variants and primers
| Gene variant | Primers |
|---|---|
| CXCR4 C-138T | 5’-AACTTCCTATGCAAGGCAGT-3’ |
| 5’-TATCTGTCATCTGCCTCACT-3’ | |
| SDF1 3’A | 5’-CAGTCAACCTGGGCAAAGCC-3’ |
| 5’-AGCTTTGGTCCTGAGAGTCC-3’ |
Table 2.
Test results for CXCR4
REFERENCES
1. Coucouvanis J, Hallas D, Farley JN. Autism Spectrum Disorder. In: Yearwood EL, Newland JA, editor. Child and Adolescent Behavioral Health. West Sussex, UK: John Wiley & Sons, Ltd, 2012, p. 238-261.
2. Casanova MF, Amaral DG, Rubensteiny JNR. Neuroscience of Autism. In: Tasman A, Kay J, Lieberman JA, First MB, Riba MB, editor. Psychiatry. 4th Ed. Chichester, UK: John Wiley & Sons, Ltd, 2015, p. 382-397.
3. Ann Le Couteur, Peter Szatmari. Autism spectrum disorder. In: Thapar A, Pine DS, Leckman JF, Scott S, Snowling MJ, Taylor E, editor. Rutter’s Child and Adolescent Psychiatry. 6th Ed. Chichester, UK: John Wiley & Sons, Ltd, 2015, p. 661-682.
4. Dembic Z. Chemokines. In: Dembic Z, editor. Cytokines of the Immune System. Amsterdam: Academic Press, 2015, p. 241-262.
5. Murdoch C, Monk PN, Finn A. Functional expression of chemokine receptor CXCR4 on human epithelial cells. Immunology 1999;98:36-41.



6. Adler MW, Geller EB, Chen X, Rogers TJ. Viewing chemokines as a third major system of communication in the brain. AAPS J 2005;7:865-870.

7. López-Bendito G, Sánchez-Alcañiz JA, Pla R, Borrell V, Picó E, Valdeolmillos M, et al. Chemokine signaling controls intracortical migration and final distribution of GABAergic interneurons. J Neurosci 2008;28:1613-1624.



8. Banisadr G, Fontanges P, Haour F, Kitabgi P, Rostène W, Mélik Parsadaniantz S. Neuroanatomical distribution of CXCR4 in adult rat brain and its localization in cholinergic and dopaminergic neurons. Eur J Neurosci 2002;16:1661-1671.


9. Ijsbrand Kramer M. Chemokines and Traffic of White Blood Cells. In: Ijsbrand Kramer M, editor. Signal Transduction. 3rd Ed. Boston: Academic Press, 2016, p. 777-812.
10. Krakowiak P, Goines PE, Tancredi DJ, Ashwood P, Hansen RL, Hertz-Picciotto I, et al. Neonatal cytokine profiles associated with autism spectrum disorder. Biol Psychiatry 2017;81:442-451.


11. Zerbo O, Yoshida C, Grether JK, Van de Water J, Ashwood P, Delorenze GN, et al. Neonatal cytokines and chemokines and risk of Autism Spectrum Disorder: the Early Markers for Autism (EMA) study: a case-control study. J Neuroinflammation 2014;11:113



12. Cash-Padgett T, Sawa A, Jaaro-Peled H. Increased stereotypy in conditional Cxcr4 knockout mice. Neurosci Res 2016;105:75-79.


13. Uppal N, Hof PR. A Stereologic Perspective on Autism Neuropathology. In: Mouton PR, editor. Neurostereology: Unbiased Stereology of Neural Systems. Ames, USA: John Wiley & Sons, Inc, 2014, p. 237-256.
14. Betancur C, Mitchell KJ. Synaptic Disorders. In: Mitchell KJ, editor. The Genetics of Neurodevelopmental Disorders. Hoboken, NJ, USA: John Wiley & Sons, Inc, 2015, p. 195-238.
15. Tucker ES, Maynard TM, LaMantia AS. Cxcr4 regulation of interneuron migration is disrupted in 22q11.2 deletion syndrome. Proc Natl Acad Sci US 2012;109:18601-18606.

16. Shi MD, Chen JH, Sung HT, Lee JS, Tsai LY, Lin HH. CXCL12-G801A polymorphism modulates risk of colorectal cancer in Taiwan. Arch Med Sci 2013;9:999-1005.



17. Okuyama Kishima M, Brajão de Oliveira K, Ariza CB, de Oliveira CE, Losi uembarovski R, Banin Hirata BK, et al. Genetic polymorphism and expression of CXCR4 in breast cancer. Anal Cell Pathol (Amst) 2015;2015:289510



19. Bacon KB, Harrison JK. Chemokines and their receptors in neurobiology: perspectives in physiology and homeostasis. J Neuroimmunol 2000;104:92-97.


20. Jiang Y, Salafranca MN, Adhikari S, Xia Y, Feng L, Sonntag MK, et al. Chemokine receptor expression in cultured glia and rat experimental allergic encephalomyelitis. J Neuroimmunol 1998;86:1-12.


21. Siebert H, Sachse A, Kuziel WA, Maeda N, Brück W. The chemokine receptor CCR2 is involved in macrophage recruitment to the injured peripheral nervous system. J Neuroimmunol 2000;110:177-185.


22. Ma Q, Jones D, Borghesani PR, Segal RA, Nagasawa T, Kishimoto T, et al. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc Natl Acad Sci USA 1998;95:9448-9453.



23. Arimitsu N, Shimizu J, Fujiwara N, Takai K, Takada E, Kono T, et al. Role of SDF1/CXCR4 interaction in experimental hemiplegic models with neural cell transplantation. Int J Mol Sci 2012;13:2636-2649.



24. Madison JM, Zhou F, Nigam A, Hussain A, Barker DD, Nehme R, et al. Characterization of bipolar disorder patient-specific induced pluripotent stem cells from a family reveals neurodevelopmental and mRNA expression abnormalities. Mol Psychiatry 2015;20:703-717.



26. Suzuki K, Matsuzaki H, Iwata K, Kameno Y, Shimmura C, Kawai S, et al. Plasma cytokine profiles in subjects with high-functioning autism spectrum disorders. PLoS ONE 2011;6:e20470



27. Abdallah MW, Larsen N, Grove J, Bonefeld-Jørgensen EC, Nørgaard-Pedersen B, Hougaard DM, et al. Neonatal chemokine levels and risk of autism spectrum disorders: findings from a Danish historic birth cohort follow-up study. Cytokine 2013;61:370-376.


28. Ashwood P, Krakowiak P, Hertz-Picciotto I, Hansen R, Pessah IN, Van de Water J. Associations of impaired behaviors with elevated plasma chemokines in autism spectrum disorders. J Neuroimmunol 2011;232:196-199.


29. Işman FK, Kucukgergin C, Daşdemir S, Cakmakoglu B, Sanli O, Seckin S, et al. Association between SDF1-3’A or CXCR4 gene polymorphisms with predisposition to and clinicopathological characteristics of prostate cancer with or without metastases. Mol Biol Rep 2012;39:11073-11079.


30. Lee YL, Kuo WH, Lin CW, Chen W, Cheng WE, Chen SC, et al. Association of genetic polymorphisms of CXCL12/SDF1 gene and its receptor, CXCR4, to the susceptibility and prognosis of non-small cell lung cancer. Lung Cancer 2011;73:147-152.


31. Ide A, Kawasaki E, Abiru N, Sun F, Fukushima T, Takahashi R, et al. Stromal-cell derived factor-1 chemokine gene variant is associated with type 1 diabetes age at onset in Japanese population. Hum Immunol 2003;64:973-978.


32. Kim YJ, Park JK, Kang WS, Kim SK, Park HJ, Nam M, et al. LAMB1 polymorphism is associated with autism symptom severity in Korean autism spectrum disorder patients. Nord J Psychiatry 2015;69:594-598.


33. Schmidt RJ, Hansen RL, Hartiala J, Allayee H, Sconberg JL, Schmidt LC, et al. Selected vitamin D metabolic gene variants and risk for autism spectrum disorder in the CHARGE Study. Early Hum Dev 2015;91:483-489.



34. Sesarini CV, Costa L, Grañana N, Coto MG, Pallia RC, Argibay PF, et al. Association between GABA(A) receptor subunit polymorphisms and autism spectrum disorder (ASD). Psychiatry Res 2015;229:580-582.


35. Saxena V, Ramdas S, Ochoa CR, Wallace D, Bhide P, Kohane I. Structural, genetic, and functional signatures of disordered neuro-immunological development in autism spectrum disorder. PLoS One 2012;7:e48835



36. Shailesh H, Gupta I, Sif S, Quhtit A. Towards understanding the genetics of Autism. Front Biosci (Elite Ed) 2016;8:412-426.


37. Cho SC, Yoo HJ, Park M, Cho IH, Kim BN, Kim JW, et al. Genome-Wide association scan of korean autism spectrum disorders with language delay: a preliminary study. Psychiatry Investig 2011;8:61-66.



38. Sweeten TL, Posey DJ, McDougle CJ. High blood monocyte counts and neopterin levels in children with autistic disorder. Am J Psychiatry 2003;160:1691-1693.








