|
 |
- Search
| Psychiatry Investig > Volume 14(6); 2017 > Article |
Abstract
In this study, we evaluated the association between autism spectrum disorders (ASDs) and 10 single-nucleotide polymorphisms (SNPs) in the 5' region of the semaphorin 5A gene (SEMA5A) for 250 Korean trios including children with ASDs. Family-based association testing and haplotype analysis revealed a statistically significant association between rs194085 and multiple sociality traits with Korean ASDs in the dominant model (p<0.001, corrected p=0.035). This indicates that genetic variations in the 5' region of SEMA5A play a role in the genetic predisposition to sociality traits in Korean ASDs.
Genetic factors are thought to play important roles in the pathogenesis of autism spectrum disorders (ASDs), and several genome-wide association studies (GWAS) have evaluated large family sizes for linkage and association mapping. Weiss et al.1 suggested that the semaphorin 5A gene (SEMA5A) is an autism susceptibility gene based on the results of a GWAS of 1031 multiplex autism families.
A previous report also observed down-regulation of SEMA5A from Epstein-Barr virus-transformed B lymphocytes from idiopathic autism patients.2 In addition, SEMA5A expression was significantly lower in autistic brains than in control brains after adjusting for age at brain acquisition, post-mortem interval, ACCESSand sex.1 In this study, we investigated whether SEMA5A is genetically associated with Korean ASDs.
A total of 250 Korean ASDs family samples from the Korean Autism Genetic Consortium were used (including 173 multiplex families and 959 individuals) in this study as previous described.3 To confirm ASD diagnoses, subjects were administered the Korean versions of the Autism Diagnostic Observation Schedule45 and the Autism Diagnostic Interview-Revised (ADI-R).67 The ADI-R is designed to elicit a full range of information needed to diagnose autism and assist in assessing ASD. The components of the diagnostic algorithm of ADI-R are subdivided into 3 main domains: qualitative abnormalities in reciprocal social interaction (designated “A”), qualitative abnormalities in communication (“B”), and repetitive and restricted behaviors and interests (“C”). The Asperger Syndrome Diagnostic Scale (ASDS) is designed to identify Asperger syndrome in children aged 5-18 years and contains 50 items grouped into 5 subgroups including sociality-related subscales.8 The social responsiveness scale (SRS) is a 65-item rating scale that generates scaled scores for specific symptom domains (social awareness, social cognition, social communication, social motivation, and autistic mannerisms), as well as a total score, which serves as an index of the severity of social deficits in ASD.9 In order to test family-based associations for ASD quantitative traits for sociality, we selected 3 traits with quantitative clinical scores using tools such as the total score of subdomain A for a diagnostic algorithm in the Korean version of ADI-R, total value of the subscale for sociality in ASDS, and converted total score for SRS (Supplementary Table 1 in the online-only Data Supplement).
The study protocol was approved by the Institutional Review Board of Eulji University (IRB No. EU 11-13). Informed consent was confirmed (or waived) by the Institutional Review Board and written informed consent was obtained from the parents of all subjects.
Single-nucleotide polymorphisms (SNPs) were selected as common tagSNPs suggested by International HapMap project website (minor allele frequencies>0.1, r2=0.8 in Chinese and Japanese populations) in a 100-kb region from rs10513025, which has been reported as a marker for SNPs in a previous GWAS study.1 Nine SNPs (rs218052, rs194085, rs150630, rs390, rs160499, rs385, rs41457, rs395, and rs41463) and the rs10513025 SNP were examined in this study. The SNPs were genotyped in genomic DNA from blood using the GoldenGate™ Assay (Illumina, San Diego, CA, USA).
Family-based association tests for each SNP and haplotype were assessed using the family-based Association Test (FBAT) program package (http://www.biostat.harvard.edu/˜ fbat/default.html). To analyze the multiple correlated traits for sociality simultaneously, we apply FBAT-GEE method.
This multivariate approach can substantially reduce the power loss associated with multiple testing approaches.10 To handle multiple comparison problems, statistical significance was specified by means of Bonferroni correction.
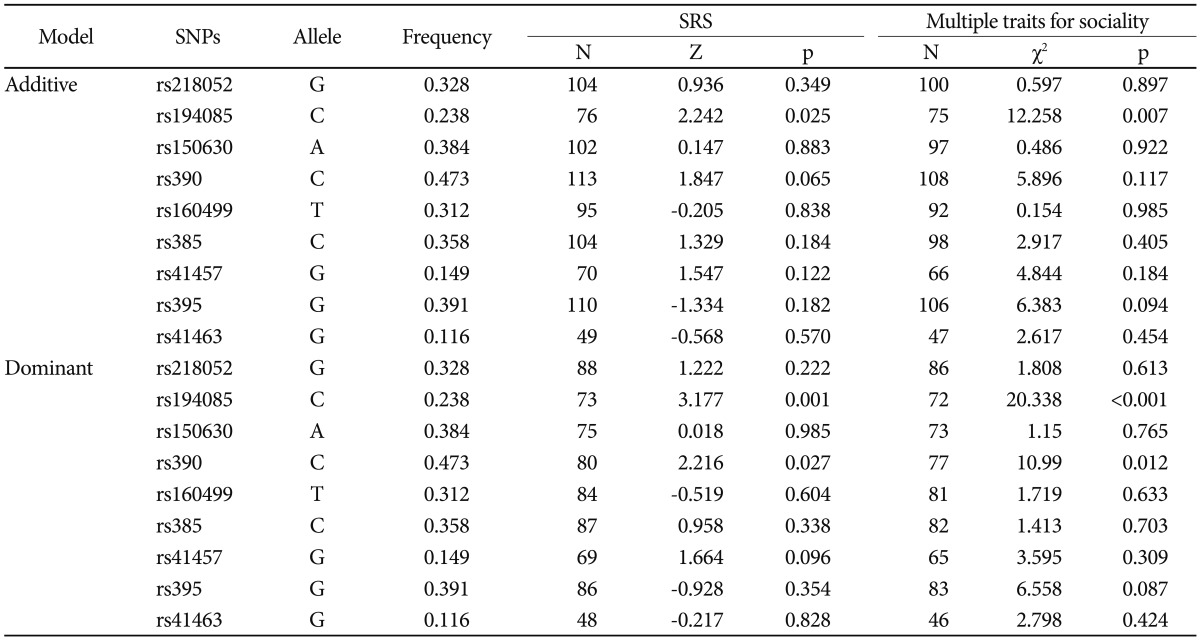
SNP rs10513025, which was reported to be associated with ASDs in a previous GWAS study, was monomorphic in this study sample. In the linkage disequilibrium (LD) test for each pair of markers, an LD block containing 3 SNPs (rs218052, rs194085, and rs150630) was observed with relatively strong LD values (0.57<D′<0.94) (Supplementary Figure 1 in the online-only Data Supplement). There was no significant difference in SNP markers for any model and any mode in FBAT for discretion affection status (Supplementary Table 2 in the online-only Data Supplement). Power was lacking to detect an association with disease affection (Supplementary Table 3 in the online-only Data Supplement). In haplotype analyses of 3 SNPs in the LD block, one haplotype showed a statistically significant association with both the additive (Z=-2.185, p=0.029) and dominant (Z=-2.041, p=0.041) models (Supplementary Table 4 in the online-only Data Supplement). However, the significance disappeared after multiple testing correction. The results of FBAT with specific traits for sociality revealed a significant difference in rs194085 with a quantitative trait for SRS-summary score in the dominant and additive models (dominant model C allele Z=3.170, p=0.001, corrected p=0.020; additive model C allele Z=2.242, p=0.025). In addition, we observed a genetic association with rs390 for the SRS score (dominant model C allele Z=2.216, p=0.027). Furthermore, when we applied the FBAT for 3 traits with additional quantitative data for sociality, the results revealed stronger associations at each SNP. Particularly, statistical significance for the genetic association with rs194085 in the dominant model remained after multiple testing correction (biallelic mode C allele Z=3.170, p<0.001, corrected p=0.005) (Table 1). rs194085 was also statistically significant in the multiallelic model (df=2, χ2=21.85, p=0.001, corrected p=0.020).
As a hypothesis-free approach, GWAS examining a sufficiently large sample and replicating the most significant SNPs in an independent sample with sufficient power is thought to avoid false-positive findings. However, GWAS cannot typically replicate previous findings. In this study, we found no association between polymorphisms in the 5′ region of SEMA5A with Korean ASDs. This may be because of a lack of power, heterogeneity across populations, technical bias, population stratification, or environmental effects.11 Anney et al.12 also failed to replicate the findings for this 5′ region of SEMA5A in 1558 autism families, likely because of a lack of power. Moreover, in contrast to the “common disease-common variant” genetic model underlying GWAS, risk alleles with low minor allele frequency or rare variants may be involved in causing ASDs. Recently, a role for rare causal variants and de novo mutations emerged in the genetic architecture of ASDs.13 In this study, common SNPs were selected, but not all polymorphisms could be examined. Ethnic differences may have affected the results because SNP rs10513025 showed a positive association with ASDs in GWAS and is monomorphic in the Korean population.1
In this study, we focused on sociality-related traits in Korean ASDs and found statistically significant associations between rs194085 SNP and sociality traits. The alleles of rs194085 showed different affinities to several transcription factors at rSNPs MAPPER2 (http://genome.ufl.edu/mapper/mapper-run?init=rs), such as the TATA box binding protein, which is thought to function as a key component of the general transcription machinery and plays a role in embryonic and nervous system development.1415 In conclusion, this study suggests that rs194085, an SNP that may affect the expression of genes involved in neurogenesis, is associated with sociality-related Korean autistic phenotypes and possibly plays a role in the pathogenesis of ASDs.
Acknowledgments
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (NRF-2014R1A2A1A11053289). Hee Jeong Yoo was supported by the Korea Healthcare Technology R&D Project, Ministry of Health & Welfare, Republic of Korea [grant number A120029]. Mira Park was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2013R1A1A2062848).
Supplementary Materials
The online-only Data Supplement is available with this article at https://doi.org/10.4306/pi.2017.14.6.876.
Supplementary Table 1
List of endophenotypes and the number of enrolled person and mean in Korean ASD patients and unaffected siblings
Supplementary Table 2
The family-based association test for ASD affection with single nucleotide polymorphisms in SEMA5A gene using biallelic mode
Supplementary Table 3
Power calculation results from TDT for discrete traits (affection) using genetic power calculator
Supplementary Table 4
Haplotype analysis for ASD affection in SEMA5A gene using FBAT
References
1. Weiss LA, Arking DE. Gene Discovery Project of Johns Hopkins & the Autism Consortium. Daly MJ, Chakravarti A. A genome-wide linkage and association scan reveals novel loci for autism. Nature 2009;462:802-808.

2. Melin M, Carlsson B, Anckarsater H, Rastam M, Betancur C, Isaksson A, et al. Constitutional downregulation of SEMA5A expression in autism. Neuropsychobiology 2006;54:64-69. PMID: 17028446.



3. Yoo HJ, Woo RS, Cho SC, Kim BN, Kim JW, Shin MS, et al. Genetic association analyses of neuregulin 1 gene polymorphism with endopheontype for sociality of Korean autism spectrum disorders family. Psychiatry Res 2015;227:366-368. PMID: 25858800.


4. Lord C, Rutter M, Goode S, Heemsbergen J, Jordan H, Mawhood L, et al. Autism diagnostic observation schedule: a standardized observation of communicative and social behavior. J Autism Dev Disord 1989;19:185-212. PMID: 2745388.


5. Yoo HJ, Kwak YS. Korean Version of Autism Diagnostic Observation Schedule (ADOS). Seoul: Hakji-sa; 2007.
6. Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord 1994;24:659-685. PMID: 7814313.


7. Yoo HJ. Korean Version of Autism Diagnostic Interview-Revised (ADI-R). Seoul: Hakji-sa; 2007.
8. Boggs KM, Gross AM, Gohm CL. Validity of the Asperger syndrome diagnostic scale. J Dev Phys Disabil 2006;18:163-182.

9. Constantino JN, Davis SA, Todd RD, Schindler MK, Gross MM, Brophy SL, et al. Validation of a brief quantitative measure of autistic traits: comparison of the social responsiveness scale with the autism diagnostic interview-revised. J Autism Dev Disord 2003;33:427-433. PMID: 12959421.


10. Lange C, Silverman EK, Xu X, Weiss ST, Laird NM. A multivariate family-based association test using generalized estimating equations: FBAT-GEE. Biostatistics 2003;4:195-206. PMID: 12925516.


11. Freitag CM, Staal W, Klauck SM, Duketis E, Waltes R. Genetics of autistic disorders: review and clinical implications. Eur Child Adolesc Psychiatry 2010;19:169-178. PMID: 19941018.


12. Anney R, Klei L, Pinto D, Regan R, Conroy J, Magalhaes TR, et al. A genome-wide scan for common alleles affecting risk for autism. Hum Mol Genet 2010;19:4072-4082. PMID: 20663923.



13. Gillis RF, Rouleau GA. The ongoing dissection of the genetic architecture of autistic spectrum disorder. Mol Autism 2011;2:12PMID: 21740537.



14. Veenstra GJ, Weeks DL, Wolffe AP. Distinct roles for TBP and TBP-like factor in early embryonic gene transcription in Xenopus. Science 2000;290:2312-2315. PMID: 11125147.


15. Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiol Rev 2009;89:121-145. PMID: 19126756.