 |
 |
- Search
| Psychiatry Investig > Volume 9(2); 2012 > Article |
Abstract
Objective
Dysfunction of neural plasticity in the brain is known to alter neural networks, resulting in depression. To understand how fluoxetine regulates molecules involved in neural plasticity, the expression levels of NCAM, NCAM140, CREB and pCREB, in rat C6 glioma cells after fluoxetine treatment were examined.
Methods
C6 cells were cultured after 20 min or after 6, 24 or 72 h treatments with 10 ┬ĄM fluoxetine. Immunocytochemistry was used to determine the effect of fluoxetine on the expression of NCAM. Western blot analysis was used to measure the expression levels of NCAM140 and CREB and the induction of pCREB after fluoxetine treatment.
Results
NCAM expression following 72-h fluoxetine treatment was significantly increased around cell membranes compared to control cells. Cells treated with fluoxetine for 6 and 72 h showed a significant increase in NCAM140 expression compared to cells treated for 20 min. The level of pCREB in the cells treated with fluoxetine for 72 h not only increased more than 60%, but was also significantly different when compared with the other treatment times. The 72-h fluoxetine treatment led to the increase of NCAM140 and the phosphorylation of CREB in C6 cells.
Recent studies have reported that dysfunction of neuronal plasticity in the brain alters neuronal networks, resulting in depression.1,2 Patients with depression show signs of morphological shrinkage, including reduced hippocampal volume, dendritic and synaptic atrophy and reduced adult neurogenesis in the brain.3-5 This shrinkage can be treated with therapeutic antidepressants, which can reverse such morphological changes through neural remodeling mechanisms, such as the expression of cytoskeletal proteins, or through cell adhesion molecules, such as neural cell adhesion molecules (NCAM).6-8 The precise mechanism by which neural remodeling and the NCAM signaling pathway are regulated by antidepressants, however, is not well-known.
NCAM, a cell adhesion molecule that is a member of the immunoglobulin superfamily, plays a significant role in neuronal development and synaptic plasticity.9,10 NCAM is also involved in learning and memory in the adult brain.9 NCAM can be modified by the addition of ╬▒-2,8-polysialic acid (PSA) and also has three major isoforms: NCAM120, NCAM140 and NCAM180. NCAM120 is an extracellular polypeptide that is attached to the cell surface, NCAM140 is located in migratory growth corns and NCAM180 is present at sites of cell-cell contact.10 NCAM activates signal transduction cascades through both homophilic and heterophilic interactions. Homophilic NCAM interactions promote the binding of p59fyn (non-receptor tyrosine kinase) and p125fak (focal adhesion kinase) and their phosphorylation, which activate the Ras-mitogen-activated protein kinase (MAPK) pathway. In turn, the cyclic-AMP response element binding protein (CREB) is subsequently phosphorylated, resulting in neurite outgrowth. Beggs et al.11 showed that NCAM140 interacted with p59fyn and p125fak, suggesting that NCAM140 regulates the FAK-Fyn-Ras-MAPK pathway through a homophilic interaction. Schmid et al.12 observed that the transfection of NCAM140 into rat B35 neuroblastoma cells resulted in NCAM140 clustering that transiently activated MAPK and induced an increase in phosphorylated CREB (pCREB). Jessen et al.13 observed that PC12-E2 cells that were transfected with a negatively interfering form of CREB did not show NCAM140-stimulated neurite outgrowth. Taken together, these data indicate that NCAM140 promotes the phosphorylation of CREB. As a result, pCREB, the activated form of CREB, is not only involved in the promotion of neurite outgrowth,10 but also in learning and long-term memory.14
Some antidepressants increase the expression of NCAM or pCREB in parts of the brain.8,15,16 Conboy et al.8 showed that agomelatine, a melatonin receptor agonist and serotonin (5-HT) receptor antagonist, induced an increase of synaptic NCAM in the rat ventral hippocampus and blocked the predator stress-induced impairment of spatial memory. Varea et al.16 observed that chronic treatment with fluoxetine increased the levels of the polysialylated form of NCAM (PSA-NCAM) in the medial prefrontal cortex but decreased its levels in the amygdale of rats. Taken together, these results indicate that antidepressants differentially regulate the expression of NCAM in different regions of the brain. The transcription factor CREB, which plays an important role in neuronal plasticity, can be phosphorylated in the rat medial prefrontal cortex and the piriform cortex by chronic treatment with the tricyclic antidepressant imipramine.15 In addition, Laifeneld et al.14 found that prolonged stress induced a decrease in pCREB in the hippocampus, prefrontal cortex and striatum of rats and that treatment with desipramine or fluoxetine induced an increase of pCREB only in the prefrontal cortex. These observations showed that these antidepressants regulate pCREB expression only in specific regions of the brain, such as the prefrontal cortex. Thus, synaptic plasticity could be improved in specific regions of the brain by the antidepressant-induced increase in NCAM and pCREB expression.
Fluoxetine, a selective 5-HT reuptake inhibitor, exhibits a strong antidepressive effect by blocking the 5-HT transporter (5-HTT), resulting in increased levels of available serotonin at the synapse.17 Despite the almost immediate effect on 5-HTT levels at the synapse after its administration, the clinical effects of fluoxetine are only seen after a few weeks of treatment.1 These delayed effects have been suggested to be associated with changes in postreceptor signaling.18,19
To better understand how fluoxetine regulates molecules involved in neuronal plasticity, this study was designed to determine the effects of time-dependent fluoxetine treatments on NCAM140 and pCREB in rat C6 glioma cells. C6 cells, which have a high level of N-methyl-D-aspartate, dopamine D2 and glucocorticoid receptors, react to antidepressants similarly in vivo and in vitro by directly down-regulating the monoamine receptor through the postsynaptic action of the antidepressants.20,21 After the treatment of C6 cells with fluoxetine, immunocytochemistry was used to examine the distribution of NCAM and western blot analysis was used to determined the expression levels of NCAM140, CREB and pCREB over time.
C6 cells (ATCC, Manassas, VA, USA) were cultured in DMEM (Invitrogen, Carlsbad, CA, USA) containing 10% FBS and penicillin (100 U/mL)/streptomycin (100 ┬Ąg/mL) (P/S) at 37Ōäā in humidified 5% CO2. For the experiments, the cells were seeded at a density of 1├Ś106 cells/100-mm culture dish. After 12 h, fresh medium containing fluoxetine (Eli Lilly Indianapolis, IN, USA) or 10% ethanol was added to the cell culture for the experimental or control cells, respectively. Ethanol was selected for the control because it was the solvent for the fluoxetine. For the fluoxetine group, the cells were incubated in fresh medium containing 10 ┬ĄM fluoxetine for 20 min or for 6, 24 or 72 h. The 10-┬ĄM concentration of fluoxetine was selected for this study because it has been shown that apoptosis does not occur under this condition.22,23 For the control group, the cells were incubated in fresh medium containing 10% ethanol for 20 min or for 6, 24 or 72 h. These treatments were repeated six times.
C6 cells were plated onto cover slips pre-coated with collagen (Sigma-Aldrich, St. Louis, MO, USA) in DMEM containing 10% FBS and P/S and incubated at 37Ōäā in humidified 5% CO2. After 12 h, fresh medium containing fluoxetine or 10% ethanol was added to the cell culture, and the cells were incubated for 24 or 72 h. After incubation, the cells were fixed in 4% paraformaldehyde for 20 min at room temperature and then incubated in PBS containing 10% normal goat serum, 0.3% Triton X-100 and 0.1% bovine serum albumin (BSA) (Sigma-Aldrich) for 30 min at room temperature. The cells were incubated with a polyclonal rabbit anti-NCAM antibody (1 : 2,000) (Chemicon, Temecula, CA, USA) overnight at 4Ōäā, followed by a wash with PBS containing 0.1% BSA. Next, the cells were incubated with biotinylated anti-rabbit IgG (H+L) (1 : 200) (Vector Laboratories, San Francisco, CA, USA) for 1 h, followed by a wash with PBS containing 0.1% BSA. The cells were then incubated with a streptavidin-conjugated anti-rabbit antibody (1 : 400) (Vector Laboratories) in the dark for 1 h. The cells were placed in a mounting solution containing 4'-6-diamidino-2-phenylindole (Vector Laboratories) and observed using a phase contrast microscope (Olympus, Tokyo, Japan).
Total protein extraction from cells was performed using the PRO-PREPŌäó Protein Extraction Solution (iNtRON Biotechnology, Seongnam, Korea) according to the manufacturer's instructions. The extracted proteins were separated by SDS-PAGE and transferred to PVDF membranes. The membranes were incubated overnight at 4Ōäā with the polyclonal rabbit anti-NCAM (1 : 10,000) (Chemicon), polyclonal rabbit anti-CREB (1 : 1,000) (Cell Signaling Technology, Danvers, MA, USA) and polyclonal rabbit anti-pCREB (1 : 1,000) (Cell Signaling Technology) antibodies, followed by incubation with HRP-conjugated anti-rabbit IgG (1 : 2,000). After washing in TBST, the reactions were visualized using ECL Plus Western Blotting Detection Reagents (Amersham Life Science, Buckinghamshire, UK). The concentration of NCAM140, CREB and pCREB was quantified using QuantityOne software (Bio-Rad Laboratories, Hercules, CA, USA).
The data were expressed as the mean ┬▒ standard error of mean (SEM) from six or more independent experiments. Statistical analyses were performed using SPSS 13.0 (SPSS Inc., Chicago, IL, USA). The data were tested using one-way ANOVAs followed by post hoc testing with Tukey's honestly significant difference (HSD) test, and p-values<0.05 were considered significant.
The effects of 24 h and 72 h fluoxetine treatment on NCAM expression in C6 cells are shown in Figure 1. The 24-h fluoxetine treatment resulted in the distribution of NCAM both around the cell membrane and in the nucleus, whereas NCAM was primarily expressed in the nucleus and around the nuclear membrane in the control cells (Figure 1A). The 72-h treatment with fluoxetine caused significantly higher levels of NCAM to be localized to the cell membranes compared to the 24-h treated cells. There were no differences with respect to NCAM expression in the control cells when 24- and 72-h cultures were compared (Figure 1B). Based on these results and the fact that NCAM140 exists in freely moving growth cones,11 the NCAM found in increased amounts around the cell membranes of 72-h fluoxetine-treated cells is likely to be NCAM140.
After the cultivation of the fluoxetine-treated cells and the 4% ethanol-treated control cells for different lengths of time (20 min and 6, 24 and 72 h), immunoblotting was used to detect the expression of NCAM140. Even after 72 h, the level of NCAM140 in the control cells did not increase (Figure 2A). In the fluoxetine-treated cells, the expression of NCAM140 increased by more than 50% at 6 and 24 h and by almost 100% at 72 h when compared to that in the cells treated with fluoxetine for 20 min (Figure 2B). In particular, cells treated with fluoxetine for 6 (p=0.044) and 72 h (p=0.003) exhibited significant increases of NCAM140 compared to cells treated for 20 min, suggesting that a 72-h treatment with fluoxetine promotes neurite outgrowth via increased levels of NCAM140. In contrast, NCAM120 expression slightly decreased, and NCAM180 expression did not change in response to fluoxetine when compared to cells treated for 20 min (Figure 3).
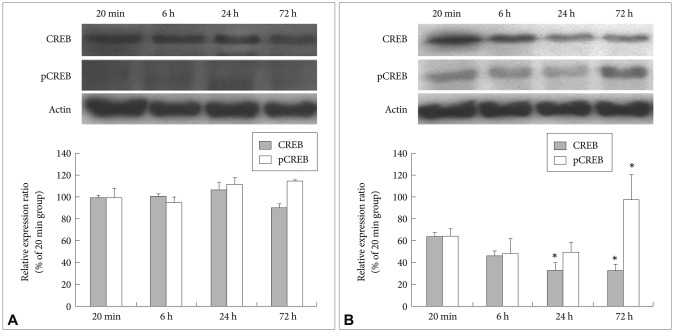
After the cultivation of the fluoxetine-treated and control cells for different lengths of time (20 min and 6, 24 and 72 h), immunoblotting was used to detect the expression and phosphorylation (pCREB) of CREB. At 72 h, the expression of CREB in the control cells was slightly increased compared to that at 20 min, although this increase was not statistically significant (Figure 4A). In addition, the levels of pCREB in the control cells increased slightly at 24 and 72 h compared to that at 20 min; however, this increase was not statistically significant, and the intensity of the pCREB bands was generally faint at all time points tested (Figure 4A). In the fluoxetine-treated cells, the expression levels of CREB continuously decreased over time, with the amount of CREB at 24 (p=0.018) and 72 h (p=0.017) significantly lower than that at 20 min (Figure 4B). In contrast, although there was a slight decrease in pCREB in the fluoxetine-treated cells at 6 and 24 h compared to that at 20 min, the levels of pCREB in the fluoxetine-treated cells at 72 h not only increased more than 60%, they were also significantly different (p<0.001) when compared to the other treatment times (Figure 4B). Taken together, these results suggest that fluoxetine promotes both the expression and phosphorylation of CREB.
In response to stress, the brains of depressive patients show glucocorticoid hypersecretion or hyperactivity of the hypothalamic- pituitary-adrenal axis, reduced hippocampal volume and dendritic remodeling. This response induces the destruction of synaptic plasticity.24 The chronic administration of antidepressants can reverse these dysfunctions.25 To understand the chronic effects of antidepressants at the molecular and cellular levels, identifying the mechanisms by which antidepressants regulate the inhibition of neuronal damage and dendrite outgrowth is important. Therefore, the aim of this study was to determine the effects of fluoxetine on the expression of NCAM140 and pCREB, which both belong to the NCAM signaling pathway that regulates neuronal remodeling.
In cells treated with fluoxetine for 24 h, the expression level of NCAM was higher in both the nucleus and cytoplasm than that in the control cells, showing the possibility that fluoxetine promoted the expression of NCAM. After a 72-h treatment with fluoxetine, NCAM was mainly localized to the cell membranes. In addition, the fluoxetine-induced increase in the levels of NCAM was time-dependent, as shown by the higher NCAM levels after the 72-h treatment compared to those after the 24-h treatment. Varea et al.16 demonstrated that in vivo chronic treatment with fluoxetine increased the levels of PSA-NCAM in the medial prefrontal cortex at approximately the same rate that the levels of NCAM increased in the present study. Chronic treatment with imipramine, a tricyclic antidepressant, had also been reported to induce an increase of PSA-NCAM in the rat prelimbic cortex.15 In contrast, Fang et al.26 reported that venlafaxine, a serotonin and norepinephrine reuptake inhibitor, attenuated the increase of NCAM in the hilus area of the mouse hippocampus. Taken together, these observations indicate that antidepressants may have different effects on NCAM expression in the brain that is region specific.
A variety of NCAMs are generated by alternative splicing, with NCAM120, NCAM140 and NCAM180 being the major splice forms. On the neuronal cell surface, NCAM140 induces the phosphorylation of p125fak and p59fyn through NCAM140 homophilic binding and subsequently activates the Ras-MAPK pathways. In turn, these two downstream effects result in an increased amount of neurite outgrowth.10 Previously, we demonstrated that chronic fluoxetine treatment promoted the mRNA expression of the NCAM140 variant in C6 cells.27 In the present study, the levels of NCAM140 protein were increased by fluoxetine treatment over time, whereas in the control cells, NCAM140 expression did not change over time. In particular, a 72-h treatment with fluoxetine resulted in an increase of NCAM140 by more than 80% when compared to a single 20-min treatment with fluoxetine. In contrast, fluoxetine did not appear to affect the levels of NCAM120 or NCAM180. Wakabayashi et al.28 demonstrated that NCAM140 mRNA levels decreased in the peripheral blood cells of bipolar disorder patients during their depressive stage, suggesting that the level of NCAM140 is involved in depression. Given that patients with major depression, bipolar disorder or schizophrenia have shown aberrant alterations of structural and synaptic plasticity such as dendritic or spine remodeling and that those dysfunctions could be normalized by antidepressants, our data suggest that the synaptic plasticity regulated by fluoxetine is controlled via the NCAMs, especially the NCAM140 isoform. Thus, fluoxetine treatment in C6 cells facilitates the expression of NCAM140 but not that of either NCAM120 or NCAM180, suggesting that fluoxetine may regulate synaptic plasticity via NCAM140.
The transcriptional activity of CREB depends on its phosphorylation state, which results in the activation of cAMP response element-mediated gene transcription and neuronal remodeling such as neurite outgrowth.29 Some studies have demonstrated that fluoxetine treatment increases the phospho-rylation of CREB.19,30 Thome et al.19 observed an increase of pCREB in several mouse brain regions, including the cerebral cortex, amygdala and hypothalamus after chronic but not acute treatment with fluoxetine. Mato et al.30 reported that chronic fluoxetine treatment induced the up-regulation of pCREB in the rat prefrontal cortex. In the present study, the 6- and 24-h treatment of C6 cells with 10 ┬ĄM of fluoxetine showed a tendency to slightly decrease the levels of pCREB, whereas the 72-h treatment induced a significant increase in pCREB expression. Fluoxetine treatment for 72 h also decreased the expression of CREB, suggesting that the effect of fluoxetine on pCREB is distinct from its effect on CREB, which may involve a negative regulatory pathway or no interaction at all. Based on previous studies19,30 and the results herein, the concentration of fluoxetine is assumed to be important with respect to its ability to increase pCREB levels, with fluoxetine treatment leading to increased pCREB both in vivo and in vitro.
In summary, fluoxetine treatment increased the levels of NCAM in rat C6 glioma cells. In particular, NCAM140, one of the NCAM major isoforms, was up-regulated by fluoxetine treatment.
Furthermore, pCREB, which is a down-stream transcription
factor in the Ras-MAPK pathway, was also increased by fluoxetine treatment. Taken together, the findings herein and those reported in previous studies12,13 suggest that fluoxetine treatment
regulates neuronal plasticity and neurite outgrowth by phosphorylating
and activating CREB via the NCAM140 homophilic
interaction-induced activation of the Ras-MAPK pathway and we illustrated the mechanism by which fluoxetine regulates the NCAM140 homophilic interaction (schematic diagram shown in Figure 5). In addition, these increases may account for the delay in therapeutic response to antidepressants at the molecular,
biological and pharmacological levels. Although the in vitro up-regulation of the NCAM140 protein by fluoxetine was described for the first time in this study, the findings reported here must be reviewed in the context of the study's limitations. Since antidepressant treatment was not performed in the serum-free conditions comparable with stressful conditions in vitro, we are unable to exclude that antidepressant might regulate other isoforms of NCAM except for NCAM140 in the serum-free conditions.
Another methodological issue is the pCREB expression
in the control cells. Although we have performed the experiments
six times to obtain pCREB immunoblot data in the control cells, the additional study could be done until they are proven to be sufficient. Therefore, further research should be performed to resolve these limitations.
References
1. Duman RS, Malberg J, Thome J. Neural plasticity to stress and antidepressant treatment. Biol Psychiatry 1999;46:1181-1191. PMID: 10560024.


3. Castren E, Rantamaki T. The role of BDNF and its receptors in depression and antidepressant drug action: reactivation of developmental plasticity. Dev Neurobiol 2010;70:289-297. PMID: 20186711.


4. Ongur D, Drevets WC, Price JL. Glial reduction in the subgenual prefrontal cortex in mood disorders. Proc Natl Acad Sci U S A 1998;95:13290-13295. PMID: 9789081.



5. Cotter D, Mackay D, Landau S, Kerwin R, Everall I. Reduced glial cell density and neuronal size in the anterior cingulate cortex in major depressive disorder. Arch Gen Psychiatry 2001;58:545-553. PMID: 11386983.


6. Gascon E, Vutskits L, Kiss JZ. Polysialic acid-neural cell adhesion molecule in brain plasticity: from synapses to integration of new neurons. Brain Res Rev 2007;56:101-118. PMID: 17658613.


7. Rutishauser U. Polysialic acid in the plasticity of the developing and adult vertebrate nervous system. Nat Rev Neurosci 2008;9:26-35. PMID: 18059411.


8. Conboy L, Tanrikut C, Zoladz PR, Campbell AM, Park CR, Gabriel C, et al. The antidepressant agomelatine blocks the adverse effects of stress on memory and enables spatial learning to rapidly increase neural cell adhesion molecule (NCAM) expression in the hippocampus of rats. Int J Neuropsychopharmacol 2009;12:329-341. PMID: 18706130.


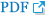
9. Luthl A, Laurent JP, Figurov A, Muller D, Schachner M. Hippocampal long-term potentiation and neural cell adhesion molecules L1 and NCAM. Nature 1994;372:777-779. PMID: 7997264.


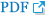
10. Povlsen GK, Ditlevsen DK, Berezin V, Bock E. Intracellular signaling by the neural cell adhesion molecule. Neurochem Res 2003;28:127-141. PMID: 12587671.


11. Beggs HE, Baragona SC, Hemperly JJ, Maness PF. NCAM140 interacts with the focal adhesion kinase p125(fak) and the SRC-related tyrosine kinase p59(fyn). J Biol Chem 1997;272:8310-8319. PMID: 9079653.


12. Schmid RS, Graff RD, Schaller MD, Chen S, Schachner M, Hemperly JJ, et al. NCAM stimulates the Ras-MAPK pathway and CREB phosphorylation in neuronal cells. J Neurobiol 1999;38:542-558. PMID: 10084688.


13. Jessen U, Novitskaya V, Pedersen N, Serup P, Berezin V, Bock E. The transcription factors CREB and c-Fos play key roles in NCAM-mediated neuritogenesis in PC12-E2 cells. J Neurochem 2001;79:1149-1160. PMID: 11752056.


14. Laifenfeld D, Karry R, Grauer E, Klein E, Ben-Shachar D. Antidepressants and prolonged stress in rats modulate CAM-L1, laminin, and pCREB, implicated in neuronal plasticity. Neurobiol Dis 2005;20:432-441. PMID: 15905095.


15. Sairanen M, O'Leary OF, Knuuttila JE, Castr├®n E. Chronic antidepressant treatment selectively increases expression of plasticity-related proteins in the hippocampus and medial prefrontal cortex of the rat. Neuroscience 2007;144:368-374. PMID: 17049169.


16. Varea E, Castillo-G├│mez E, G├│mez-Climent MA, Blasco-Ib├Ī├▒ez JM, Crespo C, Mart├Łnez-Guijarro FJ, et al. Chronic antidepressant treatment induces contrasting patterns of synaptophysin and PSA-NCAM expression in different regions of the adult rat telencephalon. Eur Neuropsychopharmacol 2007;17:546-557. PMID: 17307340.


17. Fuller RW, Wong DT, Robertson DW. Fluoxetine, a selective inhibitor of serotonin uptake. Med Res Rev 1991;11:17-34. PMID: 1994152.


18. Nibuya M, Nestler EJ, Duman RS. Chronic antidepressant administration increases the expression of cAMP response element binding protein (CREB) in rat hippocampus. J Neurosci 1996;16:2365-2372. PMID: 8601816.



19. Thome J, Sakai N, Shin K, Steffen C, Zhang YJ, Impey S, et al. cAMP response element-mediated gene transcription is upregulated by chronic antidepressant treatment. J Neurosci 2000;20:4030-4036. PMID: 10818138.



20. Yu J, Roh S, Lee JS, Yang BH, Choi MR, Chai YG, et al. The effects of venlafaxine and dexamethasone on the expression of HSP70 in rat C6 glioma cells. Psychiatry Investig 2010;7:43-48.



21. Choi MR, Oh DH, Kim SH, Yang BH, Lee JS, Choi J, et al. Fluoxetine up-regulates Bcl-xL expression in rat C6 glioma cells. Psychiatry Investig 2011;8:161-168.



22. Donati RJ, Thukral C, Rasenick MM. Chronic treatment of C6 glioma cells with antidepressant drugs results in a redistribution of Gsalpha. Mol Pharmacol 2001;59:1426-1432. PMID: 11353802.


23. Levkovitz Y, Gil-Ad I, Zeldich E, Dayag M, Weizman A. Differential induction of apoptosis by antidepressants in glioma and neuroblastoma cell lines: evidence for p-c-Jun, cytochrome c, and caspase-3 involvement. J Mol Neurosci 2005;27:29-42. PMID: 16055945.


24. McEwen BS. Effects of adverse experiences for brain structure and function. Biol Psychiatry 2000;48:721-731. PMID: 11063969.


25. Duman RS. Pathophysiology of depression: the concept of synaptic plasticity. Eur Psychiatry 2002;17(Suppl 3):306-310. PMID: 15177086.


26. Fang S, Yan B, Wang D, Bi X, Zhang Y, He J, et al. Chronic effects of venlafaxine on synaptophysin and neuronal cell adhesion molecule in the hippocampus of cerebral ischemic mice. Biochem Cell Biol 2010;88:655-663. PMID: 20651837.


27. Choi MR, Baik SY, Jung KH, Chai YG, Kim SH, Roh S, et al. Characterization of differentially expressed genes upon chronic fluoxetine treatment in rat C6 glioma cells. Korean J Psychopharmacol 2004;15:457-467.
28. Wakabayashi Y, Uchida S, Funato H, Matsubara T, Watanuki T, Otsuki K, et al. State-dependent changes in the expression levels of NCAM-140 and L1 in the peripheral blood cells of bipolar disorders, but not in the major depressive disorders. Prog Neuropsychopharmacol Biol Psychiatry 2008;32:1199-1205. PMID: 18430502.


29. D'Sa C, Duman RS. Antidepressants and neuroplasticity. Bipolar Disord 2002;4:183-194. PMID: 12180273.


30. Mato S, Vidal R, Castro E, D├Łaz A, Pazos A, Valdiz├Īn EM. Long-term fluoxetine treatment modulates cannabinoid type 1 receptor-mediated inhibition of adenylyl cyclase in the rat prefrontal cortex through 5-hydroxytryptamine 1A receptor-dependent mechanisms. Mol Pharmacol 2010;77:424-434. PMID: 19995940.


Figure┬Ā1
Effect of fluoxetine on NCAM expression in rat C6 glioma cells. Cells were treated with 10 ┬ĄM of fluoxetine for 24 or 72 h. After fixation, the cells were incubated with a polyclonal rabbit anti-NCAM antibody. A: NCAM distribution in control and fluoxetine-treated cells cultured for 24 h. B: NCAM distribution in control and fluoxetine-treated cells cultured for 72 h. Arrows show nuclei stained with DAPI. Magnification: ├Ś200. NCAM: neural cell adhesion molecules. Con: control, Flx: fluoxetine.

Figure┬Ā2
Effect of fluoxetine on NCAM140 expression in rat C6 glioma cells. Cells were treated with 10 ┬ĄM of fluoxetine for 20 min or for 6, 24 or 72 h. The expression of the NCAM140 protein was measured by western blot analysis, and actin was used as an internal control. A: Control cells. B: Fluoxetine-treated cells. The values of NCAM140 intensity are represented as the mean┬▒SEM (n=6) of the percent change from 100% of the cells cultured for 20 min. *Significantly different from 20 min by a one-way ANOVA, followed by Tukey's HSD post hoc test (*p<0.05). NCAM: neural cell adhesion molecules.

Figure┬Ā3
Effect of fluoxetine on NCAM120 and NCAM180 expression in rat C6 glioma cells. Cells were treated with 10 ┬ĄM of fluoxetine for 20 min or for 6, 24 or 72 h. The expression of the NCAM120 and NCAM180 was measured by western blot analysis, and actin was used as an internal control. A: Control cells. B: Fluoxetine-treated cells. The values of NCAM120 and NCAM180 intensity are represented as the mean┬▒SEM (n=5) of the percent change from 100% of the cells cultured for 20 min. NCAM: neural cell adhesion molecules.
Figure┬Ā4
Effect of fluoxetine on CREB and pCREB expression. Cells were treated with 10 ┬ĄM of fluoxetine for 20 min or for 6, 24 or 72 h. The expression of CREB and pCREB was measured by western blot analysis, and actin was used as an internal control. A: Control cells. B: Fluoxetine-treated cells. The values of CREB and pCREB intensity are represented as the mean┬▒SEM (n=6) of the percent change from 100% of the cells cultured for 20 min. *Significantly different from 20 min by a one-way ANOVA, followed by Tukey's HSD post hoc test (*p<0.05). CREB: cyclic-AMP response element binding protein, pCREB: phosphorylated CREB.
Figure┬Ā5
Schematic diagram showing the influence of fluoxetine on NCAM140 homophilic interaction-induced activation of Ras-MAPK pathway. This pathway is described based on the summarized western blot data and previous studies10,19,27,30 mentioned in the text. MAPK: mitogen-activated protein kinase, CREB: cAMP response element-binding.







