|
 |
- Search
| Psychiatry Investig > Volume 13(5); 2016 > Article |
Abstract
Objective
Significant progress was made in the understanding etiopathogenic factors related to MDD, including through research on the role of micro RNAs (miRs). We investigated plasma miRs as potential markers for MDD in patients treated with antidepressants.
Methods
At the initiation and at the end of twelve weeks of treatment, blood samples were collected and a structured diagnostic interview and a standardized depression rating scale for the presence and severity of major depression were done. The average decrease in HAMD score was 76.89%. Plasma miR expression profiling was performed by real time PCR. The lists of up-regulated (cut-off=2) and down-regulated miRs were imported into the miRWalk2.0 algorithm and used for target predictions. KEGG database pathways analysis was used to retrieve the pathways significantly targeted by at least two of the miRs.
Results
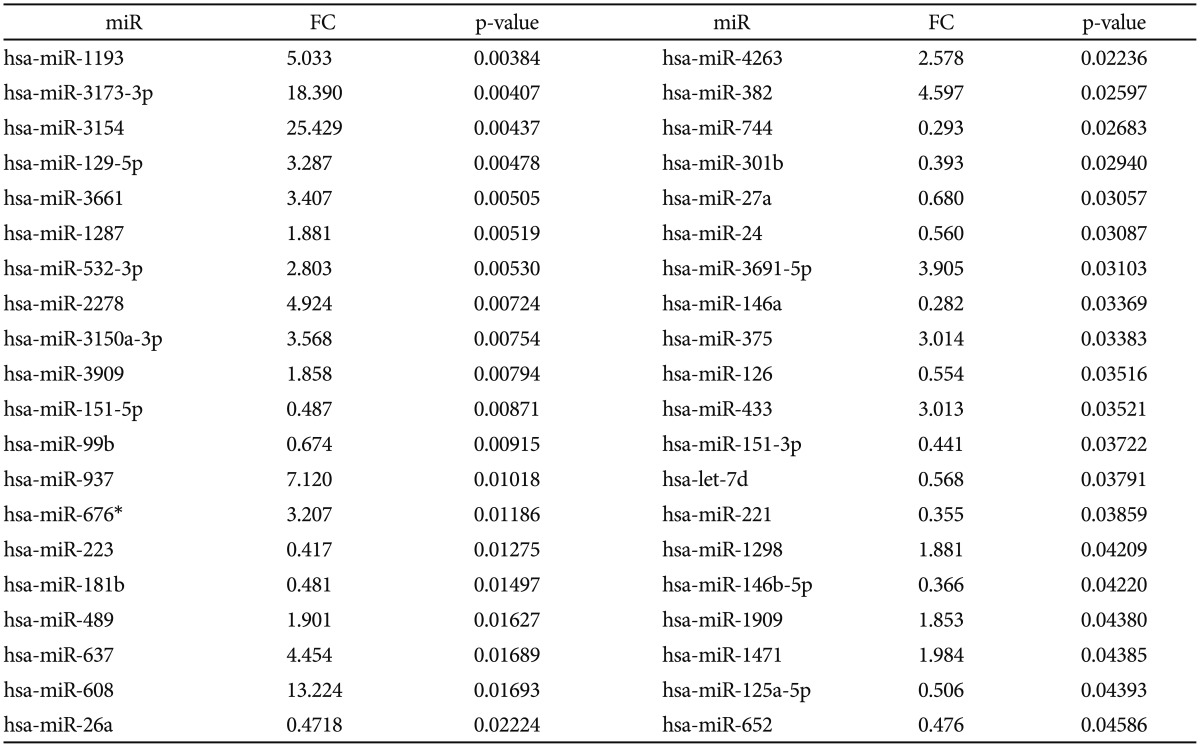
Of the 222 miRs detected in plasma samples of MDD patients, 40 were differentially expressed after treatment. Twenty-three miRs were significantly overexpressed with fold changes between 1.85 and 25.42, and 17 miRs were significantly downregulated with fold changes from 0.28 to 0.68. Pathway analysis revealed a list of 29 pathways for up-regulated miRs, and 20 pathways for down-regulated miRs. Six dysregulated miRs are common to all the top five pathways (Wnt signaling, Cancer, Endocytosis, Axon guidance, MAPK signaling): miR-146a-5p, miR-146b-5p, miR-221-3p, miR-24-3p, miR-26a-5p.
Major depression represents the most prevalent psychiatric condition existing in general population.1 Moreover, according to WHO estimates published in 1996, major depression disorder (MDD), also labeled as unipolar depression, is placed second on the list of the most disabling chronic diseases, surpassed only by cardiovascular diseases.2 To date, several distinct factors are presumed to be involved in the etiology of major depression.3 The most widely accepted etiopathogenic approach is based on the biopsychosocial model which brings into discussion several contributors that exert their long-lasting outcome starting from the antenatal period until adulthood. Moreover, the available therapeutic opportunities which mainly consist of psychological and pharmacological interventions are only partially effective, most likely due to the limitations of current medications that do not directly address the etiopathogenic determinants of MDD.4 On the other hand, it is well known that alterations of structural and neural plasticity are neurobiological markers constantly associated with MDD.5 In this respect, one could mention alterations of synaptic networks, lowering of the dorsolateral cortical activity, altering of the synaptic connectivity between the frontal lobe and other cerebral regions, modifications in the shape and number of dendritic pins, primary location of synapse formation, anomalies of the hippocampus neurons dendrites morphology, lowering of the length and shape of apical dendrites, neural atrophy and diminished hippocampus volume, decrease in the number of neurons and glial cells in cortical areas, etc.678910111213141516
It is noteworthy that recent years have marked a significant progress in the understanding of etiopathogenic factors related to MDD, including through research on the role of micro RNAs (miRs) as master regulators of gene expression. Micro RNAs are a class of small non-coding RNA molecules, 18ŌłÆ24 nucleotides long, which regulate the stability of mRNA targets and are involved in a variety of physiological and pathological conditions, including MDD and suicidal behavior.1718 In this respect, it has been shown that miR expression profiles are tissue-specific (the cerebral tissue being characterized by a complex and distinctive pattern of miRs) and differ between normal and pathological conditions,1920 MiRs are involved in multiple neurobiological processes such as neural plasticity, neurogenesis, synaptogenesis and stress response, all of these having a major role in rendering vulnerability towards MDD.212223 Recently it has been shown that certain miRs are downregulated in the prefrontal cortex of people dead by suicide and thus might associate with suicidal behavior.24 Although the research regarding miRs in MDD is still in its infancy, it is already well known that miRs play an important role in several stress related disorders including depression, as mentioned above.
The intrinsic importance of this new field of study is not only related to the discovery and validation of miRs as biomarkers for MDD, but also resides in the fact that they can be considered as new therapeutic targets for developing new classes of drugs such as anti-miRs or miR mimics. In this respect, a recent study using postmortem human brain samples has revealed that miR-1202 is specific to primates and humans and is differentially expressed in individuals with MDD, this miR regulating the gene coding for the glutamate metabotropic receptor-4 (GRM4) and can act as a predictor of therapeutic response to antidepressants from the initial phases of therapy.25 In addition, other studies on animal models have shown the utility of miRs as markers of therapeutic response to antidepressants.2627
In recent years, the presence and profile of miRs circulating in blood has been extensively studied as possible biomarkers in several pathologies, being well known that these are stable and can be accurately detected in blood and other body fluids.2829 Regarding circulating miRs as markers of MDD, research is still in its infancy and the data is currently very limited, only four such studies being published so far.30313233 The results of these studies are extremely encouraging; however they have major limitations, in the sense that, with the exception of Wan et al. (who profiled the miRs in the serum) they investigated the expression of miRs in whole blood including all blood cells. While it is questionable whether miR expression of these blood cells is representative of brain cells, it is well known that human neural tissue and specific cerebral regions have distinct miR expression profiles.
Here we report on a study investigating circulating plasma miR profiles as potential markers for MDD in patients treated with antidepressants.
All the subjects of this study are patients of the "Eduard Pamfil" Psychiatric Clinic of Timi┼¤oara who fulfilled the DSM-IV-TR diagnostic criteria for major depressive episode.34 The sampling method was one by convenience. The study was carried out in concordance with the Code of Ethics of the Declaration of Helsinki and was approved by our institutional review board. All participants provided signed informed consent.
The research is a follow-up study with two time points; the initiation of antidepressant treatment and the end of twelve weeks of antidepressant treatment. At both points we collected blood samples, and a structured diagnostic interview for major depression was done together with a standardized depression rating scale for the presence and severity of major depression.
The inclusion criteria for eligible patients were: a positive diagnostic of MDD according to DSM-IV-TR criteria (eligible patients have also undergone a SCID-I Romanian research version of clinical interview for psychiatric disorders of axis I DSM-IV-TR),35 age between 18 and 65 years old; willingness to provide an informed consent. The exclusion criteria were: having another developing psychiatric disorder diagnosed according to axe I of DSM-IV-TR; an antidepressant treatment during the last 12 months preceding the enrolment into the study; the presence of pregnancy during the study; the presence of a chronic drug treatment for a comorbid medical disease 12 months prior to or during the study. All subjects considered for the study underwent a general physical examination and a blood sample was sent to a clinical laboratory for complete blood count analysis. In order to be included in the study, subjects had to have leucocyte numbers within normal range and to be free of any sign of inflammation.
Once the diagnostic of major depressive episode was validated by the administration of SCID-I clinical interview for psychiatric disorders of axis I DSM-IV-TR, the severity of depression was subsequently assessed by using the Hamilton Depression Rating Scale (HDRS) 17-items version (also known as HAM-D). Scoring of HDRS varies depending on the version used. Hence, a score of 0ŌłÆ7 is usually accepted as being in normal range (or clinical remission) while a score of 20 or higher (indicating at least moderate severity) is usually required for inclusion in a clinical study.3637 A cut-off Ōēż7 was used to define the clinical remission of major depressive episode.
Initially, we recruited 10 subjects that respected all the aforementioned inclusion criteria, of which 2 dropped out voluntarily and 3 have not reached clinical remission after twelve weeks of treatment with escitalopram. Therefore we finally processed for miRNA analysis the plasma samples of 5 subjects, two females and 3 males with an average age of 47.20┬▒7.92 (mean┬▒standard deviation). The average decrease in HAMD score for the 5 subjects was 76.89% (HAMD score at T0=23.80┬▒2.39, HAMD score at T12=5.50┬▒1.14, for a mean dose of 12.3┬▒3.3 mg escitalopram/day, as presented in Table 1.
Blood samples were collected by venipuncture in EDTA coated vacutainers and plasma was separated by centrifugation within 2 hours of collection and stored at -80Ōäā until further use. Before RNA extraction, plasma samples were centrifuged for 5 min at 15000 g in order to eliminate cellular debris and possible cellular contamination. Total RNA containing micro RNA was extracted from 200 uL plasma using the miRNeasy Serum/Plasma kit (Qiagen), adding C. elegans miR-39 miRNA mimic spike-in as internal normalization control, according to the manufacturer's recommendations. RNA yields were assessed using a NanoDrop Spectrophotometer (Thermo Fisher Scientific) and 9 uL total RNA were used for reverse transcription with the miScript II RT Kit (Qiagen) following the manufacturer's instructions. Mature miRNA expression was determined by real time PCR in an ABI 7900HT System (Life Technologies) using the miScript miRNA PCR Array Human miRNome (Qiagen) which interrogates 1008 unique human miRNAs, following the manufacturer's procedures. Briefly, 25 uL of diluted cDNA were mixed with 1375 uL 2XQuantiTect SYBR Green PCR Master Mix (Qiagen), 275 uL 10XmiScript Universal Primer (Qiagen) and 1075 uL RNAse free water, and 25 uL of the mix were added in each well of the 96-well plates, sealed, centrifuged briefly to eliminate bubbles and then subjected to 40 cycles of real time PCR. Data were analyzed by the ╬ö╬öCT method of relative quantification, normalized to the C. elegans miR-39 miRNA spike-in as previously described.38
Only miRs showing expression in all samples were selected for further analysis. The fold changes in normalized miRNA expression between the two time points (before and after treatment) were calculated and a paired t-test was used for statistical significance.
The miR list was divided in two sublists, up-regulated (cut-off=2) and down-regulated were imported as such into the miRWalk2.0 algorithm (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/)39 and used for 3'UTR target predictions (minimum seed length 7, p-value=0.05). KEGG (Kyoto Encyclopedia of Genes and Genomes) database pathways analysis was used (with Bonferroni correction, p<0.05) to retrieve the pathways significantly targeted by at least two of the miRs.
Of all analyzed miRs, 222 were detected in all plasma samples of MDD patients in our study. Of these, 40 miRs (Escitalopram-miRs, or E-miRs) were differentially expressed after treatment, as presented in Table 2. Twenty-three miRs were significantly overexpressed with fold changes ranging between 1.85 and 25.42, the majority (17) with a fold change greater than 2.5. The rest of 17 miRs were significantly downregulated after treatment with fold changes ranging from 0.28 to 0.68.
KEGG pathways analysis of the miRs dis-regulated after Escitalopram treatment (E-miRs) using the miRWalk algorithm revealed a list of 43 pathways (at p<0.05) for up-regulated miRs, out of which 29 are targeted by at least 2 miRs. In the case of down-regulated miRs, 46 pathways (at p<0.05) out of which 20 are targeted by at least two miRs were detected. Most of these pathways (65.5% and 51.4%, for up- and down-regulated miRs, respectively) are related (directly or indirectly) to cancer (Table 3). The detailed list of the miRWalk predictions are presented in Supplementary Table 1 (in the online-only Data Supplement).
We have next asked which of the E-miRs have been already documented to target the top five pathways: Pathways in cancer, Wnt signaling, Endocytosis, Axon guidance and MAPK signaling. A miRWalk analysis of the Gene-miRNA-Pathway Interactions for all the five signaling pathways followed by a cross-examination with our list of E-miRS led to a number of 15 (Wnt signaling), 17 (Cancer), 18 (Endocytosis), 13 (Axon guidance), and 13 (MAPK signaling) already documented in the literature to target (Table 4). Six of these E-miRs are common to all the five pathways: hsa-miR-146a-5p, hsa-miR-146b-5p, hsa-miR-221-3p, hsa-miR-24-3p, hsa-miR-26a-5p. The entire list of documented E-miRS including their molecular targets and links to the literature can be found in Supplementary Table 2 (in the online-only Data Supplement).
We have investigated herein the change in expression of circulatory micro RNAs in MDD patients after 3 months of antidepressant treatment. To our knowledge, this is the first study to report changes in plasma miRNAs in MDD patients; previous studies investigating whole blood or blood white cells, only one other study analyzed miRNAs in serum.30313233 We identified 40 miRNAs with altered expression, 23 being significantly up- and 17 being down-regulated, respectively. Several of these were also found disregulated in the blood of MDD patients as discussed below.
Bocchio-Chiavetto et al.,33 determined the expression profiles of 755 miRs in the whole blood of 10 MDD patients treated with escitalopram for 3 months, showing that 30 miRs were differentially expressed after treatment, 28 being upregulated and 2 being downregulated. Only two differentially expressed miRs overlapped between this study and ours, albeit in a different direction, namely miR-26a and let-7d. Both miRs were downregulated in plasma of MDD patients after treatment in our study; however Bocchio-Chiavetto et al. reported an upregulation of these in whole blood. This difference could possibly be due to the different sample type between the studies.
Another study by Belzeaux et al.32 determined miR profiles of 762 miRs in peripheral blood mononuclear cells (PBMCs) of 13 healthy subjects and 16 MDD patients before and 2 and 8 weeks after treatment, showing different miR profiled between cases and controls, and whose expression varies according to treatment. One of the miRs (miR-433) reported in this study to be upregulated after 8 weeks of treatment in blood cells was also found to be upregulated in plasma of MDD patients after 12 weeks of treatment in our study. In addition, another miR (miR-652) found to be upregulated in blood cells of MDD patients compared to healthy controls by Blezeaux et al. was also found to be dysregulated in plasma after treatment in our study, being downregulated. This could suggest that the higher levels of miR-652 in MDD patients were lowered by antidepressant medication as found in our study.
Fan et al.30 investigated by microarray the expression of 723 miRs in PBMCs of MDD patients compared to healthy controls and fund 26 significantly differentially expressed miRs. Among the ones found at higher levels in PBMCs of MDD patients, miR-146b-5p was also found in our study to be downregulated in plasma of MDD patients after treatment, suggesting that antidepressant medication lowers the level of this miR to values closer to those found in healthy people.
In a study of 179 miRs in the cerebrospinal fluid (CSF) of 6 MDD patients compared to 6 healthy controls, Wan et al.31 reported 11 upregulated and 5 downregulated miRs, of which 3 and 1, respectively, were conformed in the serum of same subjects. Three of these miRs (miR-125a-5p, miR-221, let-7d) upregulated in the CSF and two of which confirmed in serum (miR-221, let-7d) of MDD patients were also found downregulated after treatment in plasma in our study, suggesting a normalizing effect of antidepressant treatment on the circulating miR levels. However, one other miR (miR-375) upregulated in CSF of MDD patients was also found upregulated by treatment in our study, although not confirmed in serum by Wan et al.
Several of the miRs found to be dysregulated by treatment in our study were also described as being involved in the pathology of MDD by others using human brain tissue or animal models. For example Smalheiser et al.40 reported several miRs down-regulated and reorganized in the prefrontal cortex of depressed suicide subjects, among which miR-489, miR-27a and miR-146a were also found dysregulated by treatment in our study. Moreover, downregulated miR-24 and miR-221 from our study were also found to be decreased in the hippocampus of rats after long term mood stabilizer treatment such as valproate and lithium.41 In a study of mice subjected to acute stress, Rinaldi et al.42 found increased expression in the prefrontal cortex of miR-151-3p and miR-125a-5p, which were also found downregulated by treatment in the plasma of our MDD patients. Notably, miR-125a-5p was also found at higher levels in the CSF of MDD patients as compared to healthy controls as mentioned above.31
Others have found some of the same circulating miRs as reported by us to be dysregulated in other psychiatric disorders. For example, miR-151-3p that was found at higher levels in the brain of stressed mice was also found downregulated in the serum of autistic children,43 together with miR-433 which is also upregulated by antidepressant treatment in blood cells,32 miR-489 which is also downregulated in the brain of depressed subjects,40 and miR-181b which was also found in higher levels in the serum of schizophrenia patients.44
Our KEGG pathways analysis using the miRWalk algorithm showed that the majority of the targeted pathways for both up- and down-regulated miRs relate to cancer processes. This is of particular interest since the subject of the possible interactions of antidepressants with antineoplastic therapy at the level of CYP450s has become increasingly important lately.45 Given that Escitalopram has been found to have a particularly low CYP450s inhibitory potential, our data suggest that circulating microRNAs might represent another level of interaction between selective serotonin reuptake inhibitors (SSRI) and cancer pathways. Of note, hsa-miR-151-3p is one of the 6 microRNAs found to be altered by another SSRI (paroxetine) in human lymphoblastoid cell lines derived from healthy adult female individuals,46 while hsa-miR-221 is one of the microRNAs shown to be down-regulated following chronic paroxetine exposure of LCLs.47
Two major signaling pathways stand out as major targets of the escitalopram-dependent microRNAs: Wnt and MAPK signaling, both being known to be involved in depression through cell proliferation and plasticity of the hippocampus.4849 The link between SSRI and Wnt/GSK-3╬▓/╬▓-catenin signaling has been demonstrated in the dorsal hippocampus, where GSK-3╬▓ was found to be activated by citalopram.50 Our data indicate that SSRI in general, and Escitalopram in particular might modulate Wnt and MAPK signaling pathways through an epigenetic mechanism involving altered levels of circulating microRNAs.
Our miRWalk analysis shows that both up- and down-regulated miRs target the endocytosis pathway; an interesting results since it has been postulated that the trafficking of cellular serotonin transporters from and towards the cell membrane might be regulated at posttranscriptional level.51
Axon growth was also among the top pathways targeted by the escitalopram-miRs, a result reminding of previous ex vivo and in vivo studies on embryonic thalamic axons, effect which, interestingly, was described as SERT independent.52
Interestingly, none of the circulating microRNAs targets the metabotropic glutamate-receptor 4 previously shown to be the target of the primate-specific, brain enriched miR-1202 found to be involved in major depression and antidepressant treatment.25
Overall, our miRWalk analysis of the extensive changes in plasma microRNAs after Escitalopram treatment of patients diagnosed with major depression might opens a new avenue for the understanding of Escitalopram mode of action but also for its side effects.
Despite being a pilot study of small sample size, with no validation of individual miRs, to our knowledge this is the first study to assess circulating extracellular levels of miRs affected by antidepressant treatment in MDD patients, also investigating an unprecedented number of miRs; the other two existing studies examining miRs in whole blood and PBMCs, respectively.
We believe that this field of study is one of great importance and relevance, taking into account the fact that current studies have suggested a possible role of miRs as diagnostic and prognostic markers in MDD, also opening new avenues for therapeutic discoveries with a more specific action compared to existing treatments.
Supplementary Materials
The online-only Data Supplement is available with this article at http://dx.doi.org/10.4306/pi.2016.13.5.549.
References
1. Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry 2005;62:593-602. PMID: 15939837.


2. Murray CJL, Lopez AD. Alternative projections of mortality and disability by cause 1990-2020: Global Burden of Disease Study. Lancet 1997;349:1498-1504. PMID: 9167458.


3. Laursen TM, Munk-Olsen T, Nordentoft M, Bo Mortensen P. A comparison of selected risk factors for unipolar depressive disorder, bipolar affective disorder, schizoaffective disorder, and schizophrenia from a danish population-based cohort. J Clin Psychiatry 2007;68:1673-1681. PMID: 18052560.


4. Thase ME, Ninan PT, Musgnung JJ, Trivedi MH. Remission with venlafaxine extended release or selective serotonin reuptake inhibitors in depressed patients: a randomized, open-label study. Prim Care Companion CNS Disord 2011;13:PCC.10m00979PMID: 21731835.



5. Ota KT, Duman RS. Environmental and pharmacological modulations of cellular plasticity: role in the pathophysiology and treatment of depression. Neurobiol Dis 2013;57:28-37. PMID: 22691453.


6. Aganova EA, Uranova NA. Morphometric analysis of synaptic contacts in the anterior limbic cortex in the endogenous psychoses. Neurosci Behav Physiol 1992;22:59-65. PMID: 1614620.


7. Dolan RJ, Bench CJ, Liddle PF, Friston KJ, Frith CD, Grasby PM, et al. Dorsolateral prefrontal cortex dysfunction in the major psychoses; symptom or disease specificity? J Neurol Neurosurg Psychiatry 1993;56:1290-1294. PMID: 8270929.



8. Drevets WC, Ong├╝r D, Price JL. Reduced glucose metabolism in the subgenual prefrontal cortex in unipolar depression. Mol Psychiatry 1998;3:190-191. PMID: 9672886.


9. Andreasen NC. Linking mind and brain in the study of mental illnesses: a project for a scientific psychopathology. Science 1997;275:1586-1593. PMID: 9054346.


10. Honer WG. Assessing the machinery of mind: synapses in neuropsychiatric disorders. J Psychiatry Neurosci 1999;24:116-121. PMID: 10212553.


11. Toni N, Buchs PA, Nikonenko I, Bron CR, Muller D. LTP promotes formation of multiple spine synapses between a single axon terminal and a dendrite. Nature 1999;402:421-425. PMID: 10586883.


12. Hajsza T, MacLusky NJ, Leranth C. Short-term treatment with the antidepressant fluoxetine triggers pyramidal dendritic spine synapse formation in rat hippocampus. Eur J Neurosci 2005;21:1299-1303. PMID: 15813939.


13. McEwen BS. Effects of adverse experiences for brain structure and function. Biol Psychiatry 2000;48:721-731. PMID: 11063969.


14. Sheline YI. 3D MRI studies of neuroanatomic changes in unipolar major depression: the role of stress and medical comorbidity. Biol Psychiatry 2000;48:791-800. PMID: 11063975.


15. Sala M, Perez J, Soloff P, Ucelli di Nemi S, Caverzasi E, Soares JC, et al. Stress and hippocampal abnormalities in psychiatric disorders. Eur Neuropsychopharmacol 2004;14:393-405. PMID: 15336301.


16. Rajkowska G, Miguel-Hidalgo JJ. Gliogenesis and glial pathology in depression. CNS Neurol Disord Drug Targets 2007;6:219-233. PMID: 17511618.



17. Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol 2008;9:219-230. PMID: 18270516.


18. Serafini G, Pompili M, Hansen KF, Obrietan K, Dwivedi Y, Shomron N, et al. The involvement of microRNAs in major depression, suicidal behavior, and related disorders: a focus on miR-185 and miR-491-3p. Cell Mol Neurobiol 2014;34:17-30. PMID: 24213247.


19. Barry G. Integrating the roles of long and small non-coding RNA in brain function and disease. Mol Psychiatry 2014;19:410-416. PMID: 24468823.


20. Adlakha YK, Saini N. Brain microRNAs and insights into biological functions and therapeutic potential of brain enriched miRNA-128. Mol Cancer 2014;13:33PMID: 24555688.



21. Dwivedi Y. Emerging role of microRNAs in major depressive disorder: diagnosis and therapeutic implications. Dialogues Clin Neurosci 2014;16:43-61. PMID: 24733970.



22. Im HI, Kenny PJ. MicroRNAs in neuronal function and dysfunction. Trends Neurosci 2012;35:325-334. PMID: 22436491.



23. Malphettes L, Fussenegger M. Impact of RNA interference on gene networks. Metab Eng 2006;8:672-683. PMID: 16996764.


24. Smalheiser NR, Lugli G, Rizavi HS, Torvik VI, Turecki G, Dwivedi Y. MicroRNA expression is down-regulated and reorganized in prefrontal cortex of depressed suicide subjects. PLoS One 2012;7:e33201PMID: 22427989.



25. Lopez JP, Lim R, Cruceanu C, Crapper L, Fasano C, Labonte B, et al. miR-1202 is a primate-specific and brain-enriched microRNA involved in major depression and antidepressant treatment. Nat Med 2014;20:764-768. PMID: 24908571.



26. Baudry A, Mouillet-Richard S, Schneider B, Launay JM, Kellermann O. miR-16 targets the serotonin transporter: a new facet for adaptive responses to antidepressants. Science 2010;329:1537-1541. PMID: 20847275.


27. O'Connor RM, Grenham S, Dinan TG, Cryan JF. microRNAs as novel antidepressant targets: converging effects of ketamine and electroconvulsive shock therapy in the rat hippocampus. Int J Neuropsychopharmacol 2013;16:1885-1892. PMID: 23684180.


28. Witwer KW. Circulating MicroRNA Biomarker Studies: Pitfalls and Potential Solutions. Clin Chem 2015;61:56-63. PMID: 25391989.


29. Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem 2010;56:1733-1741. PMID: 20847327.



30. Fan HM, Sun XY, Guo W, Zhong AF, Niu W, Zhao L, et al. Differential expression of microRNA in peripheral blood mononuclear cells as specific biomarker for major depressive disorder patients. J Psychiatr Res 2014;59:45-52. PMID: 25201637.


31. Wan Y, Liu Y, Wang X, Wu J, Liu K, Zhou J, et al. Identification of differential microRNAs in cerebrospinal fluid and serum of patients with major depressive disorder. PLoS One 2015;10:e0121975PMID: 25763923.



32. Belzeaux R, Bergon A, Jeanjean V, Loriod B, Formisano-Tr├®ziny C, Verrier L, et al. Responder and nonresponder patients exhibit different peripheral transcriptional signatures during major depressive episode. Transl Psychiatry 2012;2:e185PMID: 23149449.



33. Bocchio-Chiavetto L, Maffioletti E, Bettinsoli P, Giovannini C, Bignotti S, Tardito D, et al. Blood microRNA changes in depressed patients during antidepressant treatment. Eur Neuropsychopharmacol 2013;23:602-611. PMID: 22925464.


34. American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4th Ed, Text Revision. Washington DC: American Psychiatric Association; 2000.
35. First MB, Spitzer RL, Gibon M, Williams JBW. Structured Clinical Interview for DSM-IV-TR Axis I Disorders, Research version (SCID-I RV). New York: Biometrics Research; 2002.
36. Hamilton M. Development of a rating scale for primary depressive illness. Br J Soc Clin Psychol 1967;6:278-296. PMID: 6080235.


37. Lam RW, Michalak EE, Swinson RP. Assessment Scales in Depression, Mania and Anxiety. London, UK: Taylor and Francis; 2005.
38. Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 2010;50:298-301. PMID: 20146939.



39. Dweep H, Sticht C, Pandey P, Gretz N. miRWalk - database: prediction of possible miRNA binding sites by ŌĆ£walkingŌĆØ the genes of 3 genomes. J Biomed Inform 2011;44:839-847. PMID: 21605702.


40. Smalheiser NR, Lugli G, Rizavi HS, Torvik VI, Turecki G, Dwivedi Y. MicroRNA expression is down-regulated and reorganized in prefrontal cortex of depressed suicide subjects. PLoS One 2012;7:e33201PMID: 22427989.



41. Zhou R, Yuan P, Wang Y, Hunsberger JG, Elkahloun A, Wei Y, et al. Evidence for selective microRNAs and their effectors as common long-term targets for the actions of mood stabilizers. Neuropsychopharmacology 2009;34:1395-1405. PMID: 18704095.


42. Rinaldi A, Vincenti S, De Vito F, Bozzoni I, Oliverio A, Presutti C, et al. Stress induces region specific alterations in microRNAs expression in mice. Behav Brain Res 2010;208:265-269. PMID: 19913057.


43. Mundalil Vasu M, Anitha A, Thanseem I, Suzuki K, Yamada K, Takahashi T, et al. Serum microRNA profiles in children with autism. Mol Autism 2014;5:40PMID: 25126405.



44. Shi W, Du J, Qi Y, Liang G, Wang T, Li S, et al. Aberrant expression of serum miRNAs in schizophrenia. J Psychiatr Res 2012;46:198-204. PMID: 22094284.


45. Miguel C, Albuquerque E. Drug interaction in psycho-oncology: antidepressants and antineoplastics. Pharmacology 2011;88:333-339. PMID: 22123153.


46. Oved K, Morag A, Pasmanik-Chor M, Oron-Karni V, Shomron N, Rehavi M, et al. Genome-wide miRNA expression profiling of human lymphoblastoid cell lines identifies tentative SSRI antidepressant response biomarkers. Pharmacogenomics 2012;13:1129-1139. PMID: 22909203.


47. Oved K, Morag A, Pasmanik-Chor M, Rehavi M, Shomron N, Gurwitz D. Genome-wide expression profiling of human lymphoblastoid cell lines implicates integrin beta-3 in the mode of action of antidepressants. Transl Psychiatry 2013;3:e313PMID: 24129413.



48. Duric V, Banasr M, Licznerski P, Schmidt HD, Stockmeier CA, Simen AA, et al. A negative regulator of MAP kinase causes depressive behavior. Nat Med 2010;16:1328-1332. PMID: 20953200.



49. Karege F, Perroud N, Burkhardt S, Fernandez R, Ballmann E, La Harpe R, et al. Protein levels of ╬▓-catenin and activation state of glycogen synthase kinase-3╬▓ in major depression. A study with postmortem prefrontal cortex. J Affect Disord 2012;136:185-188. PMID: 22036797.


50. Liu R, Dang W, Jianting M, Su C, Wang H, Chen Y, et al. Citalopram alleviates chronic stress induced depression-like behaviors in rats by activating GSK3╬▓ signaling in dorsal hippocampus. Brain Res 2012;1467:10-17. PMID: 22634067.


51. Lau T, Horschitz S, Berger S, Bartsch D, Schloss P. Antidepressant-induced internalization of the serotonin transporter in serotonergic neurons. FASEB J 2008;22:1702-1714. PMID: 18216289.


52. Bonnin A, Zhang L, Blakely RD, Levitt P. The SSRI citalopram affects fetal thalamic axon responsiveness to netrin-1 in vitro independently of SERT antagonism. Neuropsychopharmacology 2012;37:1879-1884. PMID: 22414815.



Table┬Ā3
Pathways targeted by E-miRs

E-miRs: miRs disregulated after Escitalopram treatment, No. miR: number of miRs dis-regulated in each pathway, Wnt: Wingless-related integration site, MAPK: mitogen-activated protein kinases, ErbB: ErbB receptor tyrosine kinases, mTOR: mammalian target of rampycin, VEGF: vascular endothelial growth factor, GnRH: gonadotropin releasing hormone, TGF: tumor growth factor