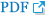|
 |
- Search
| Psychiatry Investig > Volume 17(9); 2020 > Article |
|
Abstract
Objective
Schizophrenia is one of the most devastating neuropsychiatric disorders. Genetic epidemiological studies have confirmed that schizophrenia is a genetic disease. Genes promoting neurodevelopment may be potential candidates for schizophrenia. As an adaptor linking a number of tyrosine kinase receptors in multiple intracellular signaling cascades, Src homology 2 domain containing transforming protein 3 (SHC3) is a member of the Shc-like adaptor protein family, and expressed predominantly in the mature neurons of the central nervous system (CNS). In the present study, we aimed to investigate the association of SHC3 and schizophrenia.
Methods
An independent case-control association study was performed in a sample including 710 schizophrenia patients and 1314 healthy controls from a Northeast Chinese Han population.
Schizophrenia is one of the most devastating neuropsychiatric disorders with a lifetime prevalence of about 1% worldwide. It is characterized by a breakdown in cognition and emotion along with characteristic positive (i.e., hallucinations, delusions, or disorganized speech and thoughts) and negative (i.e., apathy, alogia, or avolition) symptoms [1,2]. The disease usually starts in young adults, with a prolonged course of illness and a high disability rate, placing a heavy economic burden on individuals, families and society. So far, the etiology and pathogenesis of schizophrenia are unknown yet. It is generally believed that biological, psychological and social environmental factors and their interactions may be related to the onset of schizophrenia [3].
Genetic epidemiological studies have confirmed that schizophrenia is a genetic disease with heritability estimates of approximately 80% [4]. The pedigree survey showed that schizophrenia has familial clustering: the incidence of close relatives is higher than that of the general population. Futhermore, the closer the relationship is, the higher the risk of the disease. At the same time, twin research found that the same rate of schizophrenia among identical twins was 4-6 times higher than that of fraternal twins. Although schizophrenia shows a clear genetic tendency, classic Mendelian inheritance cannot explain the genetic laws of the disease, which means schizophrenia is a multi-gene complex disease caused by the synergy of multiple minor genes [5].
With the advancement of molecular biology techniques and methods for obtaining genetic data, the screening and research of candidate genes for schizophrenia have entered a period of rapid development. Candidate genes for schizophrenia are genes that have obtained some evidence that they may be associated with schizophrenia. Src homology 2 domain-containing transforming protein 3 (SHC3), also known as N-Shc, ShcC or Rai, is a member of the Shc-like adaptor protein family, which serves to link a number of tyrosine kinase receptors with multiple intracellular signaling cascades [6]. SHC3 protein is expressed predominantly in the mature neurons of the central nervous system (CNS), especially in striatum and rest of the CNS grey matter [7].
Previous study have shown that SHC3 as a critical hub for neurotrophic pathways control of neuronal metabolism and functions in the adulthood, such as Alzheimer disease and other metabolic and degeneratibe CNS diseases, like ParkinsonŌĆÖs disease and glaucoma [8-11]. Apart from that, some study suggested that SHC3 plays an important role in the regulation of NMDA receptor function in hippocampus and affect learning and memory [12]. Moreover, ectopic or abnormal SHC3 expression has been associated to some types of cancer, including aggressive neuroblastomas and human ependymomas [13,14]. SHC3 is also involved in Phosphoinositide 3-kinase (PI3K)-Akt signaling pathway and interacts with sphingosine-1-phosphate receptor 3 [15,16]. The roles of SHC3 in human brain may contribute to the symptomatology of patients with schizophrenia [17]. In the present study, an independent casecontrol analysis was carried out to investigate the association of SHC3 polymorphisms and schizophrenia in a Northeast Chinese Han population.
Subjects were all recruited in the Northeast China from Department of Psychiatry, Dalian Seventh PeopleŌĆÖs Hospital, P.R. China, and unrelated Chinese Han nationality born. The sample set included 710 schizophrenia patients (334 males and 376 females; mean age: 29.92┬▒9.95 years) and 1,314 healthy controls (658 males and 656 females; mean age: 27.90┬▒9.39 years). The consensus diagnoses were made by at least two experienced psychiatrists according to the diagnostic and Statistical Manual of Mental Disorders, fourth edition criteria [18] on the basis of clinical observation, medical records, and family information. The patients were diagnosed with different subtypes of schizophrenia (85.9% paranoid, 9.0% undifferentiated, 2.1% disorganized, 0.4% residual, and 2.5% undefined). All healthy controls in this study were selected by a simple non-structured interview without any Diagnostic and Statistical Manual of Mental Disorders, fourth edition axis I disorder and matched for age, sex, education, and ethnicity to the patients. The objectives and procedures of the study were explained to all participants and written informed consent was obtained. The study was approved by the Ethics Committee of the Dalian Seventh PeopleŌĆÖs Hospital, P.R. China (2015-11).
The SNPs were selected by downloading the information of all the SNPs within and neighboring the human SHC3 gene from the International HapMap project database on dbSNP (http://www.ncbi.nlm.nih.gov/SNP/). The total coverage of the 9 selected SNPs was approximately 15 kb.
Nine single nucleotide polymorphisms (SNPs) in the human SHC3 gene region were selected. Genomic DNA was extracted from venous blood by using a commercially QIAamp DNA Blood Mini Kit (QIAGEN, Hilden, German). All the SNPs were genotyped by either polymerase chain reaction (PCR) restriction fragment length polymorphism analysis or direct DNA sequencing. All pairs of PCR primers were designed by software Primer Premier 5.0 (Premier, Palo Alto, CA, USA). PCR products were either completely digested with 4 U of restriction enzyme overnight and then separated by agarose gel electrophoresis (2-3% gel) stained with ethidium bromide or sequenced on an ABI PRISM 377-96 DNA Sequencer (Applied Biosystems, Foster City, CA, USA) after purifying them using a BigDye Terminator Cycle Sequencing Ready Reaction Kit. All the results were read by two experienced technicians independently.
Deviation of the genotypes from the Hardy-Weinberg equilibrium was examined by a Žć2 goodness-of-fit test (Table 1). Distribution of gender and the difference of age between cases and controls were evaluated by Pearson Žć2-test and StudentŌĆÖs t-test with the Statistical Package for Social Science (SPSS Inc., Chicago, IL, USA) 17.0. Statistical differences in allelic and genotypic distribution between patients and controls were evaluated by the Pearson Žć2-test at a significance level of 0.05. The haplotype frequencies were estimated by the expectation maximization algorithm. Odds ratio (OR) and their 95% confidence intervals (95% CI) were calculated to evaluate the effect of different alleles and haplotypes. The analyses were performed by the Haploview version 4.1 (http://www.broad.mit.edu/mpg/haploview) [19] and SHEsis (http://analysis2.bio-x.cn/myAnalysis.php) [20,21].
According to the GTEx database, the SHC3 gene is highly expressed in brain tissue compared with other tissues (Figure 1). None of the genotype distributions of the 9 selected SNPs in case and control groups deviated from Hardy-Weinberg equilibrium. Detailed information and location of the selected SNPs were shown in Table 1. After the statistical analysis, the complete association results were listed in Table 2. Of the selected SNPs, 4 SNPs showed statistical differences in allele frequencies between cases and controls: rs2316280, p=0.016; rs4877041, p=0.016; rs7021743, p=0.0003; and rs9444485, p=0.010. After the Bonferroni correction, one of the SNPs (rs7021743, p=0.0027) remained significant.
To further investigate genotypic association of the nine SNPs with schizophrenia, we examined these SNPs under different genetic models. After analyzing the association of the SNPs by five genetic models (codominant, dominant, recessive, overdominant, and log-additive models) with schizophrenia, the consequences demonstrated that three of them showed significant differences between cases and control under at least three different genetic models, and one of them showed difference between cases and control under codominant genetic model: 1) rs2316280, codominant (p=0.021), dominant (p=0.017) and overdominant (p=0.022); 2) rs4877041, codominant (p=0.012), dominant (p=0.047), recessive (p=0.0131), and log-additive (p=0.0143); 3) rs944485, codominant (p=0.014), dominant (p=0.011), overdominant (p=0.015), and log-additive (p=0.037); and 4) rs7021743, codominant (p=0.0003) (Table 3).
To investigate whether the haplotype constructed by the four schizophrenia-associated SHC3 SNPs would result in a higher risk for schizohrpenia, global and individual haplotype associations of these four SNPs were tested (Table 4). The global associaton analysis revealed a positive result (Žć2=11.233, p=0.004). The A-A-A-G haplotype combination was different in frequency between cases and controls (Žć2=6.575, p=0.010), and the G-A-A-T haplotype combination also showed association with schizophrenia (Žć2=5.063, p=0.024).
Schizophrenia is a psychiatric syndrome characterized by positive symptoms such as hallucinations, delusions and speech disorders, negative symptoms such as decreased motivation and expression. It also presents some cognitive symptoms such as impaired executive function, memory and mental processing speed [1,2]. Schizophrenia, with a prevalence of nearly 1%, is one of the top 10 causes of disability worldwide, but daily functioning varies widely among patients, with some severely disabled and others functioning well [22]. Based on twin and family studies, genetic factors explain 80% of the risk of schizophrenia at the population level [4]. However, only a small fraction of these heritable components can be attributed to common single nucleotide variations with weak effects, and attributed to relatively large but rare mutations with relatively large effects. In genome-wide association analysis and gene expression profiling, many genes related to schizophrenia are involved in immune system, cytoskeleton development, synaptic plasticity and functional pathways. Environmental factors, including obstetric complications, early adverse experiences, and childhood residence in urban areas, may interact with genetic risk factors to influence an individualŌĆÖs susceptibility to schizophrenia [22].
SHC3 (ShcC/N-Shc/Rai), a member of the family of Shclike adaptor proteins, as a new regulator of migration of normal and cancer stem/progenitor cells [23]. The family consists of four members that collectively act as central participants in signaling pathways triggered by tyrosine kinase-coupled surface receptors, controlling various cellular processes, including proliferation, differentiation, survival, and movement [24]. There have been several lines of clinical and genetic evidence showing that SHC3 might be linked to various neurological diseases. Previous evidence demonstrated that SHC3 has been involved in the inactivation of the P13K-AKT signaling pathway [25], and SHC3 silencing may aggravate oxidative stress (OS) injury in nigral dopamine neurons via downregulation of the PI3K-AKT-FoxO signaling pathway in ParkinsonŌĆÖs disease (PD) rats [9]. Apart from that, SHC3 not only mediates trkb-ras/MAPK signaling, but also participates in the regulation of NMDA receptor function in the hippocampus, and acts as a regulator of hippocampal synaptic plasticity based on learning and memory [12]. In addition, a study demonstrated that the activation of N-Shc signaling pathway contributes to an acute kanic acid (KA)-induced epileptiform activity and neuronal cell loss in the hippocampus, which presents as a potential target for the novel therapeutic approaches of epilepsy [26].
In the present study we examined the association between the SHC3 gene and the risk of schizophrenia in a Northeast Chinese Han population. This study included 740 schizophrenia patients and 1314 healthy controls. Allelic frequencies of 4 (rs2316280, rs4877041, rs7021743, and rs9444485) out of 9 selected SNPs covering the whole SHC3 gene region showed differences between cases and controls. The genotypic association of the nine SNPs under different models by multiple logistic regression analyses showed that the four SNPs were associated with schizophrenia significantly under at least one genetic model. In addition, the haplotype constructed by these four SNPs was globally associated with schizophrenia, and two individual haplotype combinations (A-A-A-G and G-A-A-T) of these SNPs were significantly different between cases and controls. These data suggested SHC3 as a susceptibility gene for schizophrenia in a Northeast Chinese Han population.
Our study first reported that the human SHC3 gene is susceptible to schizophrenia. False positive results may occur in case-control studies due to sample size and possible population stratification. However, Japanese scientist Tomo Okochi reported that there is no association analysis between SHC3 and schizophrenia in a Japanese population at 2nd Biennial Schizophreina International Research Conference [27]. Ethnic differences may be the reason why our study is inconsistent with the conclusion of that study. Therefore, to further confirm the susceptibility of SHC3 gene to schizophrenia, repeated case-control association studies and pedigree association studies with more genetic markers and larger independent samples in different ethnic groups are needed.
In conclusion, our study indicated SHC3 gene as a susceptibility gene of schizophrenia. This study has some limitations. First, as a case-control association analysis of gene SNP, the sample size of this study is still relatively small. Second, only 9 SNPs of SHC3 gene were selected for association analysis. Therefore, in order to verify the relationship between SHC3 gene polymorphism and schizophrenia, more rigorous biological functional experiment is needed to conduct in large sample population, such as the effect of risk type functional polymorphism on disease incidence and etc.
ACKNOWLEDGEMENTS
We extend our gratitude to all the patients participating in this study. This work was supported by grants from the National Natural Science Foundation of China (81601174 and 81671298), and Natural Science Foundation of Liaoning Province, China (2019-ZD-0938). Teaching Reform Project Foundation for Undergraduate Innovative Talent Cultivation of Dalian Medical University (111806010301).
The authors have no potential conflicts of interest to disclose.
Author Contributions
Conceptualization: Zhi-Lin Luan. Data curation: Ye Lv, Yang Sun, ZhiLin Luan. Formal analysis: Ye Lv. Funding acquisition: Zhi-Lin Luan. Investigation: Yang Sun. Methodology: Zhi-Lin Luan, Ye Lv. Project administration: Yang Sun. Resources: Zhi-Lin Luan, Yang Sun. Software: Ye Lv. Supervision: Jian Yin. Validation: Guan-Yu Wang. Visualization: Cheng-Jie Li, Yi-Yang Luo. WritingŌĆöoriginal draft: Ye Lv. WritingŌĆöreview & editing: Zhi-Lin, Luan.
Figure┬Ā1.
SHC3 gene expression in different tissues. TPM on the vertical axis represents the transcript quantification value, and the horizontal axis represents different tissues. The SHC3 gene was highly expressed in brain tissue compared with other tissues.
Table┬Ā1.
List of SNPs included in the present study
| rs code | Position* | Distance from SNP1 (kb) | Allele change | HCBŌĆĀ MAF | Sample-set HWE p | Case HWE p | Control HWE p |
|---|---|---|---|---|---|---|---|
| rs3812501 | 90814217 | 0 | G>A | 0.244 | 0.407 | 0.899 | 0.382 |
| rs2316280 | 90825605 | 1.139 | G>A | 0.049 | 0.932 | 0.991 | 0.953 |
| rs4877041 | 90843569 | 2.935 | A>G | 0.024 | 0.113 | 0.006 | 0.723 |
| rs7021743 | 90856632 | 4.242 | A>C | 0.012 | 0.669 | 0.675 | 0.868 |
| rs944485 | 90869638 | 5.542 | G>T | 0.024 | 0.661 | 0.794 | 0.654 |
| rs1547696 | 90883940 | 6.972 | C>T | 0.037 | 0.998 | 0.828 | 0.944 |
| rs4876968 | 90916697 | 10.248 | G>A | 0.012 | 0.547 | 0.718 | 0.645 |
| rs1556384 | 90953947 | 13.973 | G>A | 0.024 | 0.426 | 0.579 | 0.575 |
| rs4534195 | 90958468 | 14.425 | T>C | 0.037 | 0.931 | 0.927 | 0.923 |
Table┬Ā2.
Allele frequencies of 9 SNPs in the human SHC3 gene between schizophrenia patients and controls
| rs code | Subjects | Allele and | frequency* | Žć2 p (pŌĆĀ) | OR (95% CI) |
|---|---|---|---|---|---|
| A | G | ||||
| rs3812501 | Case | 269 (0.189) | 1,151 (0.811) | Žć2=0.386 | 1.054 (0.893-1.244) |
| Control | 477 (0.182) | 2,151 (0.818) | p=0.535 | ||
| A | G | ||||
| rs2316280 | Case | 53 (0.037) | 1,367 (0.963) | Žć2=5.844 | 0.674 (0.488-0.930) |
| Control | 143 (0.054) | 2,485 (0.946) | p=0.016ŌĆĪ | ||
| A | G | ||||
| rs4877041 | Case | 1,352 (0.952) | 68 (0.048) | Žć2=5.792 | 0.673 (0.486-0.931) |
| Control | 2,542 (0.967) | 86 (0.033) | p=0.016ŌĆĪ | ||
| A | C | ||||
| rs7021743 | Case | 1,398 (0.985) | 22 (0.015) | Žć2=13.215 | 0.291 (0.144-0.591) |
| Control | 2,616 (0.995) | 12 (0.005) | p=0.0003 (0.0027)ŌĆĪ | ||
| G | T | ||||
| rs944485 | Case | 1,334 (0.939) | 86 (0.061) | Žć2=6.687 | 0.684 (0.512-0.914) |
| Control | 2,517 (0.958) | 111 (0.042) | p=0.010ŌĆĪ | ||
| C | T | ||||
| rs1547696 | Case | 1,372 (0.966) | 48 (0.034) | Žć2=0.867 | 0.840 (0.581-1.213) |
| Control | 2,553 (0.971) | 75 (0.029) | p=0.352 | ||
| A | G | ||||
| rs4876968 | Case | 19 (0.013) | 1,401 (0.987) | Žć2=0.049 | 1.066 (0.604-1.882) |
| Control | 33 (0.013) | 2,595 (0.987) | p=0.824 | ||
| A | G | ||||
| rs1556384 | Case | 29 (0.020) | 1,391 (0.980) | Žć2=1.489 | 1.349 (0.833-2.185) |
| Control | 40 (0.015) | 2,588 (0.985) | p=0.222 | ||
| C | T | ||||
| rs4534195 | Case | 51 (0.036) | 1,369 (0.964) | Žć2=1.485 | 1.251 (0.872-1.794) |
| Control | 76 (0.029) | 2,552 (0.971) | p=0.223 |
Table┬Ā3.
Genotype association of 9 SNPs in the human SHC3 gene under different genetic models with schizophrenia
| rs code | Subjets | Genotype* |
Codominant |
Dominant |
Recessive |
Overdominant |
Log-additive |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | p | OR (95%CI) | p | OR (95%CI) | p | OR (95%CI) | p | p | |||||
| AA | AG | GG | |||||||||||
| rs3812501 | Case | 26 (0.037) | 217 (0.306) | 467 (0.658) | 1.079 (0.882-1.322) | 0.456 | 1.073 (0.887-1.302) | 0.472 | 0.998 (0.623-1.598) | 0.992 | 1.001 (0.857-1.171) | 0.986 | 0.758 |
| Control | 48 (0.037) | 381 (0.290) | 885 (0.674) | 1.026 (0.638-1.652) | 0.917 | ||||||||
| AA | AG | GG | |||||||||||
| rs2316280 | Case | 1 (0.001) | 51 (0.072) | 658 (0.927) | 0.675 (0.484-0.940) | 0.021ŌĆĀ | 0.668 (0.481-0.925) | 0.017ŌĆĀ | 2.165 (0.361-26.53) | 0.479 | 1.48 (1.062-2.062) | 0.022ŌĆĀ | 0.054 |
| Control | 4 (0.003) | 135 (0.103) | 1,175 (0.894) | 2.24 (0.373-27.45) | 0.459 | ||||||||
| AA | AG | GG | |||||||||||
| rs4877041 | Case | 647 (0.911) | 58 (0.082) | 5 (0.007) | 0.762 (0.543-1.075) | 0.126 | 1.408 (1.001-1.978) | 0.047ŌĆĀ | 0.107 (0.013-0.773) | 0.0131ŌĆĀ | 0.768 (0.548-1.082) | 0.135 | 0.0143ŌĆĀ |
| Control | 1,229 (0.935) | 84 (0.064) | 1 (0.001) | 0.105 (0.012-0.758) | 0.012ŌĆĀ | ||||||||
| AA | AC | CC | |||||||||||
| rs7021743 | Case | 688 (0.969) | 22 (0.031) | 0 (0.000) | 0.024 (0.003-0.144) | 0.0003ŌĆĀ | - | - | - | - | - | - | - |
| Control | 1,302 (0.991) | 12 (0.009) | 0 (0.000) | - | - | ||||||||
| GG | GT | TT | |||||||||||
| rs944485 | Case | 627 (0.883) | 80 (0.113) | 3 (0.004) | 0.682 (0.505-0.922) | 0.014ŌĆĀ | 1.478 (1.091-2.006) | 0.011ŌĆĀ | 0.539 (0.126-2.316) | 0.443 | 0.684 (0.506-0.924) | 0.015ŌĆĀ | 0.037ŌĆĀ |
| Control | 1,206 (0.918) | 105 (0.080) | 3 (0.002) | 1.923 (0.448-8.255) | 0.8139 | ||||||||
| CC | CT | TT | |||||||||||
| rs1547696 | Case | 663 (0.934) | 46 (0.065) | 1 (0.001) | 0.849 (0.578-1.237) | 0.397 | 1.188 (0.821-1.735) | 0.371 | 0.540 (0.034-10.27) | 0.658 | 1.178 (0.808-1.730) | 0.399 | 0.064 |
| Control | 1,240 (0.944) | 73 (0.056) | 1 (0.001) | 0.5347 (0.033-10.17) | 0.653 | ||||||||
| AA | AG | GG | |||||||||||
| rs4876968 | Case | 0 (0.000) | 19 (0.027) | 691 (0.973) | 1.067 (0.606-1.882) | 0.823 | - | - | - | - | - | - | - |
| Control | 0 (0.000) | 33 (0.025) | 1,281 (0.975) | - | - | ||||||||
| AA | AG | GG | |||||||||||
| rs1556384 | Case | 0 (0.000) | 29 (0.041) | 681 (0.959) | 1.356 (0.830-2.203) | 0.218 | - | - | - | - | - | - | 0.218 |
| Control | 0 (0.000) | 40 (0.030) | 1,274 (0.970) | - | - | ||||||||
| CC | CT | TT | |||||||||||
| rs4534195 | Case | 1 (0.001) | 49 (0.069) | 660 (0.930) | 1.243 (0.850-1.802) | 0.2523 | 0.799 (0.554-.161) | 0.234 | 0.540 (0.034-10.27) | 0.658 | 1.242 (0.849-1.800) | 0.254 | 0.471 |
| Control | 1 (0.001) | 74 (0.056) | 1,239 (0.943) | 1.877 (0.099-35.69) | 0.6509 | ||||||||
Table┬Ā4.
Estimated haplotype frequencies and case-control haplotype results of the four schizophrenia-associated SNPs of human SHC3 gene
| Haplotype |
Haplotype frequency* |
Žć2 | p | OR (95% CI) |
Global |
||
|---|---|---|---|---|---|---|---|
| Case | Control | Žć2 | p | ||||
| A-A-A-G | 48.00 (0.034) | 136.74 (0.052) | 6.575 | 0.010ŌĆĀ | 0.647 (0.462-0.904) | 11.233 | 0.004ŌĆĀ |
| G-A-A-G | 1,257.08 (0.885) | 2,344.47 (0.892) | 0.522 | 0.470 | 1.099 (0.850-1.421) | ||
| G-A-A-T | 45.52 (0.032) | 54.96 (0.021) | 5.063 | 0.024ŌĆĀ | 1.574 (1.057-2.345) | ||
REFERENCES
1. Saha S, Chant D, Welham J, McGrath J. A systematic review of the prevalence of schizophrenia. PLoS Med 2005;2:e141



2. Tandon R, Keshavan MS, Nasrallah HA. Schizophrenia, ŌĆ£just the factsŌĆØ what we know in 2008. 2. Epidemiology and etiology. Schizophr Res 2008;102:1-18.


3. Ross CA, Margolis RL, Reading SA, Pletnikov M, Coyle JT. Neurobiology of schizophrenia. Neuron 2006;52:139-153.


4. Cardno AG, Marshall EJ, Coid B, Macdonald AM, Ribchester TR, Davies NJ, et al. Heritability estimates for psychotic disorders: the Maudsley twin psychosis series. Arch Gen Psychiatry 1999;56:162-168.


5. International Schizophrenia Consortium; Purcell SM, Wray NR, Stone JL, Visscher PM, OŌĆÖDonovan MC, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009;460:748-752.



6. OŌĆÖBryan JP, Songyang Z, Cantley L, Der CJ, Pawson T. A mammalian adaptor protein with conserved Src homology 2 and phosphotyrosine-binding domains is related to Shc and is specifically expressed in the brain. Proc Natl Acad Sci U S A 1996;93:2729-2734.



7. Ponti G, Conti L, Cataudella T, Zuccato C, Magrassi L, Rossi F, et al. Comparative expression profiles of ShcB and ShcC phosphotyrosine adapter molecules in the adult brain. Neuroscience 2005;133:105-115.


8. Triaca V, Sposato V, Bolasco G, Ciotti MT, Pelicci P, Bruni AC, et al. NGF controls APP cleavage by downregulating APP phosphorylation at Thr668: relevance for AlzheimerŌĆÖs disease. Aging Cell 2016;15:661-672.



9. Gong J, Zhang L, Zhang Q, Li X, Xia XJ, Liu YY, et al. Lentiviral VectorMediated SHC3 silencing exacerbates oxidative stress injury in nigral dopamine neurons by regulating the PI3K-AKT-FoxO signaling pathway in rats with ParkinsonŌĆÖs disease. Cell Physiol Biochem 2018;49:971-984.


10. Xie Z, Dong Y, Maeda U, Xia W, Tanzi RE. RNA interference silencing of the adaptor molecules ShcC and Fe65 differentially affect amyloid precursor protein processing and Abeta generation. J Biol Chem 2007;282:4318-4325.


11. Tarr PE, Roncarati R, Pelicci G, Pelicci PG, DŌĆÖAdamio L. Tyrosine phosphorylation of the beta-amyloid precursor protein cytoplasmic tail promotes interaction with Shc. J Biol Chem 2002;277:16798-16804.


12. Miyamoto Y, Chen L, Sato M, Sokabe M, Nabeshima T, Pawson T, et al. Hippocampal synaptic modulation by the phosphotyrosine adapter protein ShcC/N-Shc via interaction with the NMDA receptor. J Neurosci 2005;25:1826-1835.



13. Magrassi L, Marziliano N, Inzani F, Cassini P, Chiaranda I, Skrap M, et al. EDG3 and SHC3 on chromosome 9q22 are co-amplified in human ependymomas. Cancer Lett 2010;290:36-42.


14. Miyake I, Ohira M, Nakagawara A, Sakai R. Distinct role of ShcC docking protein in the differentiation of neuroblastoma. Oncogene 2009;28:662-673.


15. Magrassi L, Conti L, Lanterna A, Zuccato C, Marchionni M, Cassini P, et al. Shc3 affects human high-grade astrocytomas survival. Oncogene 2005;24:5198-5206.


16. Conti L, Sipione S, Magrassi L, Bonfanti L, Rigamonti D, Pettirossi V, et al. Shc signaling in differentiating neural progenitor cells. Nature Neurosci 2001;4:597-586.


17. Wong J, Rothmond DA, Webster MJ, Weickert CS. Increases in two truncated TrkB isoforms in the prefrontal cortex of people with schizophrenia. Schizophr Bull 2013;39:130-140.


18. American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders (4th Ed Text Revision). Washington: American Psychiatric Publishing; 2000.
19. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263-265.


20. Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res 2005;15:97-98.


21. Li Z, Zhang Z, He Z, Tang W, Li T, Zeng Z, et al. A partition-ligationcombination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res 2009;19:519-523.


23. Ortensi B, Osti D, Pellegatta S, Pisati F, Brescia P, Fornasari L, et al. Rai is a new regulator of neural progenitor migration and glioblastoma invasion. Stem Cells 2012;30:817-832.


24. Finetti F SM, Baldari CT. Positive and negative regulation of antigen receptor signaling by the Shc family of protein adapters. Inmmun Rev 2009;232:115-134.

25. Azzalin A, Moretti E, Arbustini E, Magrassi L. Cell density modulates SHC3 expression and survival of human glioblastoma cells through Fak activation. J Neurooncol 2014;120:245-256.