 |
 |
- Search
| Psychiatry Investig > Volume 20(12); 2023 > Article |
|
Abstract
Objective
Methods
Results
Notes
Availability of Data and Material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
Ki Woong Kim, a contributing editor of the Psychiatry Investigation, was not involved in the editorial evaluation or decision to publish this article. J.B.B., S.L., J.S.K., J.W.H., and K.W.K. received royalty income from VUNO Inc.
H.O, J.S., and D.L. are employees at VUNO Inc. All remaining authors have declared no conflicts of interest.
Author Contributions
Conceptualization: Jong Bin Bae, Subin Lee, Hyunwoo Oh, Jinkyeong Sung. Data curation: Jong Bin Bae, Subin Lee. Formal analysis: Jong Bin Bae, Subin Lee. Funding acquisition: Ki Woong Kim. Investigation: Jong Bin Bae, Subin Lee. Methodology: Jong Bin Bae, Subin Lee, Hyunwoo Oh. Project administration: Jun Sung Kim, Ji Won Han. Resources: Ji Won Han, Jae Hyoung Kim, Sang Eun Kim. Software: Hyunwoo Oh, Dongsoo Lee. Supervision: Ki Woong Kim. Validation: Jong Bin Bae, Subin Lee. Visualization: Subin Lee, Dongsoo Lee. WritingŌĆöoriginal draft: Jong Bin Bae, Subin Lee. WritingŌĆöreview & editing: all authors.
Funding Statement
This study was supported by a grant from the Korean Health Technology R&D Project, Ministry for Health, Welfare, and Family Affairs, Republic of Korea [grant number HI09C1379 (A092077)] and the Institute for Information & Communications Technology Promotion (IITP) grant funded by the Korean government (MSIT) (2018-0-00861, Intelligent SW Technology Development for Medical Data Analysis).
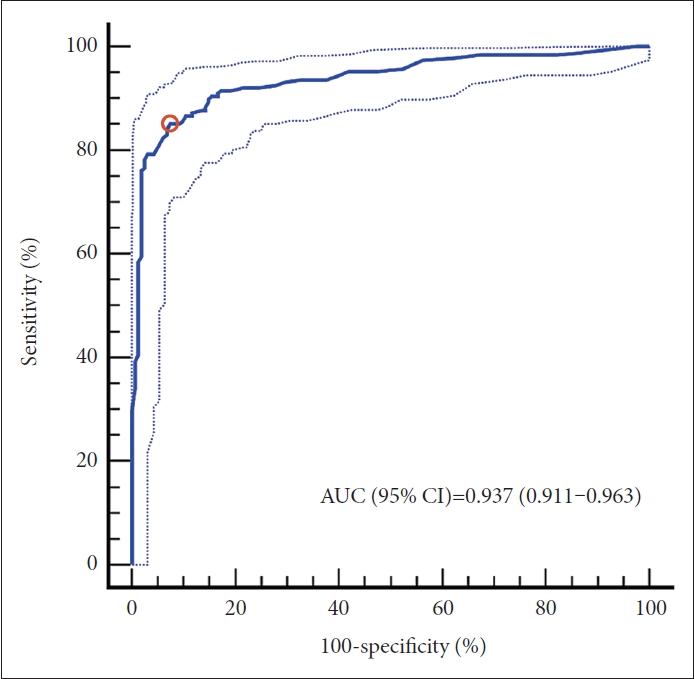
Figure┬Ā1.
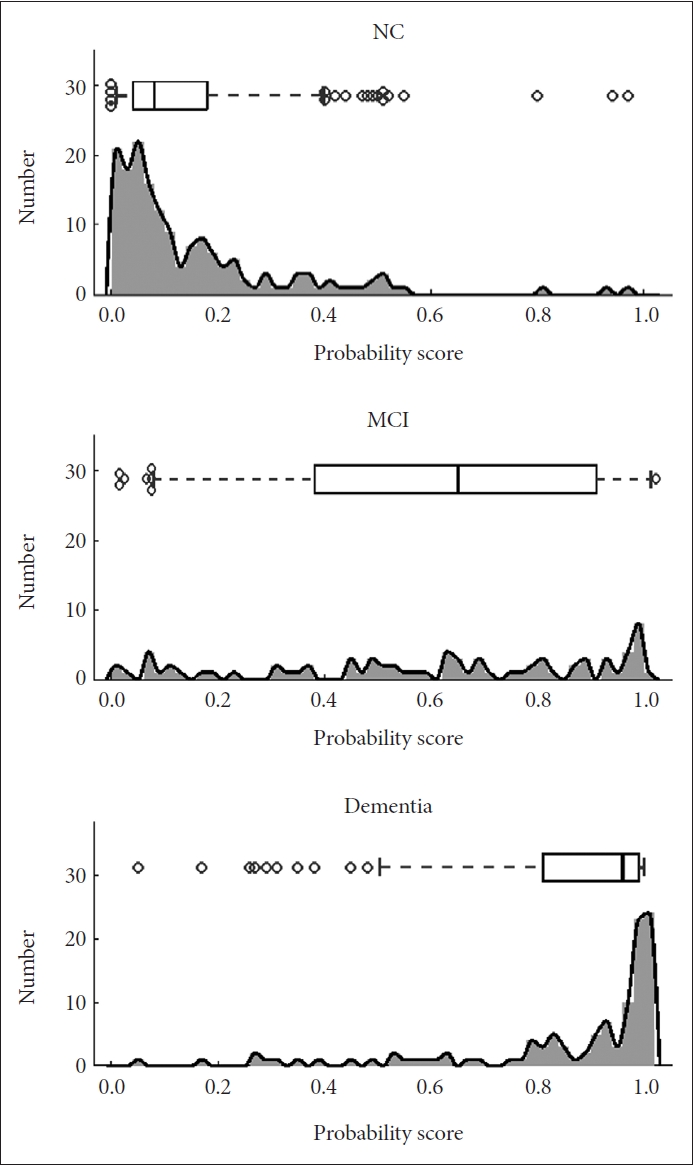
Figure┬Ā2.
Table┬Ā1.
| Characteristics | NC (N=162)* | AD (N=188)ŌĆĀ | t or Žć2ŌĆĪ | pŌĆĪ |
|---|---|---|---|---|
| Age (yr) | 73.3┬▒6.9 | 73.9┬▒7.4 | -0.8 | 0.42 |
| Age band | 13.650 | 0.009 | ||
| ŌĆā50-59 yr | 0 (0.0) | 12 (6.4) | ||
| ŌĆā60-69 yr | 46 (28.4) | 36 (19.1) | ||
| ŌĆā70-79 yr | 84 (51.9) | 96 (51.1) | ||
| ŌĆā80-89 yr | 32 (19.7) | 43 (22.9) | ||
| ŌĆāŌēź90 yr | 0 (0.0) | 1 (0.5) | ||
| Female | 108 (66.6) | 125 (66.5) | 0.001 | 0.97 |
| Education (yr) | 12.4┬▒4.5 | 11.1┬▒4.9 | 2.57 | 0.01 |
| MMSE (score) | 27.5┬▒2.2 | 20.9┬▒4.9 | 16.21 | <0.001 |
| MRI | ||||
| ŌĆāScanner | 1.64 | 0.44 | ||
| ŌĆāŌĆāPhilips Achieva | 137 (84.6) | 167 (88.8) | ||
| ŌĆāŌĆāPhilips Ingenia | 20 (12.3) | 18 (9.6) | ||
| ŌĆāŌĆāPhilips Ingenia CX | 5 (3.1) | 3 (1.6) | ||
| ŌĆāHead coil | 63.79 | <0.001 | ||
| ŌĆāŌĆāSENSE-Head-8 | 73 (45.1) | 39 (20.7) | ||
| ŌĆāŌĆāSENSE-NV-16 | 15 (9.3) | 89 (47.4) | ||
| ŌĆāŌĆāDual coil | 42 (25.9) | 39 (20.7) | ||
| ŌĆāŌĆāMulti coil | 32 (19.7) | 21 (11.2) | ||
| WMH (cc) | 14.3┬▒29.6 | 16.0┬▒17.5 | -0.64 | 0.52 |
Table┬Ā2.
Table┬Ā3.
REFERENCES







