|
 |
- Search
| Psychiatry Investig > Volume 11(3); 2014 > Article |
Abstract
Objective
Autism spectrum disorders (ASDs) are a group of early childhood-onset neurodevelopmental disorders characterized by deficits in social interaction and language skills, and repetitive behaviors. Brain-derived neurotrophic factor (BDNF) plays a critical role in the differentiation of normal neuronal cells during embryonic and postnatal neuronal development through its neurotrophic effects.
Methods
In this study, we performed a family-based association test (FBAT) between single nucleotide polymorphisms (SNPs; rs6265, rs11030101, rs7103411, and rs7103873) or haplotypes in the BDNF gene and affection status or several quantitative traits characterized by ADI-R with151 Korean trios, including a child diagnosed as ASDs.
Results
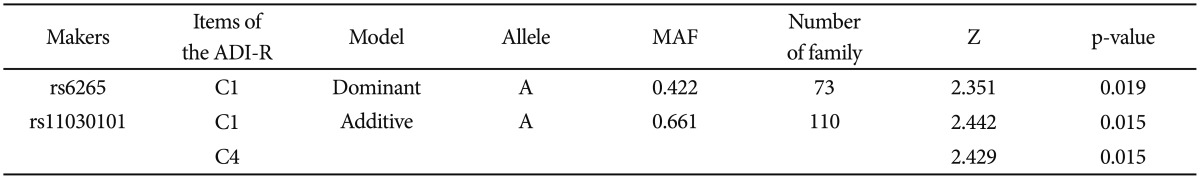
While no significant association was found between SNPs or haplotypes and the ASDs disease status, a quantitative transmission disequilibrium test (QTDT) by using quantitative traits identified associations of the SNPs (rs6265 and rs11030101) with a domain score for "Restricted, Repetitive and Stereotyped patterns of behavior" (C domain), especially at the subdomain scores for "encompassing preoccupation or circumscribed pattern of interest" (C1) (rs6265A allele, dominant model, p-value=0.019; rs11030101 A allele, additive model, p-value=0.015) and "preoccupations with part of objects or non-functional elements of material" (C4) (rs11030101 A allele, additive model, p-value=0.015) within the ADI-R diagnostic algorithm. In addition, significant associations were also identified between the haplotypes and these quantitative traits (C1, p-value=0.016; C4, p-value=0.012).
Autism spectrum disorders (ASDs) are severe neurodevelopmental disorders accompanied by dysfunctions, including communication, social interaction, restricted and repetitive behaviors, and intellectual disability with multiple undefined etiologies.1 The high heritability of ASDs has been well documented, and is reflected in the increased risk of recurrence in families and a concordance rate of 70-90% in monozygotic twins.2 Despite the evidence for a genetic basis of ASDs, the heterogeneity of previous genetic association studies has hampered the evaluation genes of involved in ASD phenotypes,3 and it has been suggested that multiple genetic factors are implicated in the pathogenesis of ASDs.
Brain-derived neurotrophic factor (BDNF), a member of the neurotrophin family, is a prosurvival factor that plays a key role in regulating neuronal survival, formation of functional synapses, plasticity of synaptic connections, and modulation of the differentiation and maintenance of phenotype in mature neurons throughout the human brain.4,5 Studies of BDNF levels in the plasma, serum, and brain of autistic subjects have shown a possible association between increased BDNF levels and ASDs,6,7 and the modulation of BDNF level in these neurological disorders has been suggested as a potential therapeutic approach.8
The Val66Met (rs6265) BDNF gene polymorphism is a well-documented functional SNP. This SNP has been implicated in anxiety-related personality traits and in the pathogenesis of conditions accompanied by anxiety and depression, such obsessive-compulsive disorder and attention-deficit hyperactivity disorder (ADHD), which are thought to be phenotypic variations in ASDs.9 Moreover, a relationship has also been shown between the BDNF Val66Met genotype and the regional cortical surface area of ASD patients.10 Furthermore, Han et al.11 reported an association between BDNF haploinsufficiency and low adaptive behavior and reduced cognitive function in WAGR/11p13 deletion syndrome. In addition, in a genetic association study of the BDNF gene with ASDs, Nishimura et al.12 reported a family-based association between ASDs and haplotypes containing the BDNF gene rs11030121 SNPs.
The aims of this study were to investigate family-based association between 4 SNPs of the BDNF gene and ASDs in a Korean population and to evaluate the relationship between BDNF genotypes and clinical phenotypes characterized by a diagnostic algorithm for ASDs.
All 151 families in the study consisted of a trio (3 individuals: the father, mother, and child); signed informed consent was obtained from either the parents or caregivers, and the study method was confirmed and approved by the Institutional Review Board of Euji University. Descriptions of the ASD subject selection and diagnostic procedures have been previously published by our group.13 Briefly, children with ASDs were clinically evaluated using the DSM-VI diagnostic criteria intelligence and social maturity, and then confirmed using the Korean versions of the Autism Diagnostic Observation Schedule (K-ADOS) and Autism Diagnostic Interview-Revised (K-ADI-R).14,15 Subjects with organic brain disease, chromosomal aneuploidy, and tuberous sclerosis were ruled out. The probands that met the ASD diagnostic criteria comprised 86.1% male, 87.4% autistic disorder, 13.5% PDD-NOS, and 1.6% Asperger's syndrome. DNA was extracted from peripheral blood samples of all individuals in the 151 families by using a G-spin Genomic DNA Extraction Kit (Intron, Daejeon, Korea).
The human BDNF gene (NM_170731) is located between nucleotide positions 27,633,018 and 27,677,756 in chromosome 11p14.1 and contains 2 exons (UCSC Genome Browser GRCh36/hg18, http://genome.ucsc.edu). Candidate SNPs in the BDNF were evaluated using publicly available genotype data from Tokyo (JPT) populations by using the International HapMap project (www.hapmap.org, HapMap data release 27 Phase II+III) and the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/snp, Entrez SNP Database). The nonsynonymous SNP rs6265 (Val66Met) has been well defined in a variety of psychiatric diseases and has a major allele frequency (MAF) of 0.344 for the A allele in JPT. The SNPs rs11030101, rs7103411, and rs7103873 in intron 1 were selected using Tag SNP picker (www.hapmap.org) and genotyped using the GoldenGateŌäó Assay (Illumina, San Diego, CA, USA).
Hardy-Weinberg equilibrium (HWE) and linkage disequilibrium (LD) between pairs of SNPs were calculated using Haploview 3.2.16 Prior to analyses, Mendelian errors were verified using family-based association test (FBAT) from the FBAT package.17 FBATs were conducted by using the additive, dominant, and recessive models. The HBAT application in the FBAT package was used to test the associations between haplotypes and ASDs. The HBAT calculation was computed using the Monte Carlo option with 10,000 replications.
Of the 151 subjects, 6 were omitted from analysis of the relationship between susceptible alleles and phenotypes. For the analyses of association with genotypic polymorphism, the mean of sub-domain scores was compared in each genotype group. The test for association between the specific genotype of polymorphisms and the 3 ADI-R domain scores (qualitative abnormalities in reciprocal social interaction [A]; qualitative abnormalities in communication [B]; and restricted, repetitive, and stereotyped patterns of behavior [C]) was conducted with quantitative trait analysis in FBAT. The B domain was classified for both verbal and nonverbal subjects, and then the sum of scores for the groups was calculated. The C domain was divided into the following sub-domains: C1 (encompassing preoccupation or circumscribed pattern of interest), C2 (apparent compulsive adherence to nonfunctional routines or rituals), and C4 (preoccupations with part of objects or non-functional elements of material). Based on the diagnostic algorithm of ADI-R, the scores of the ADI-R items were as follows: 0 (no abnormality) to 3 (most abnormality), and responses such as "not applicable" or "unknown or not asked" were considered missing data.18 A previous study has shown the ADI-R subdomains.13 Statistical significance was set at p<0.05.
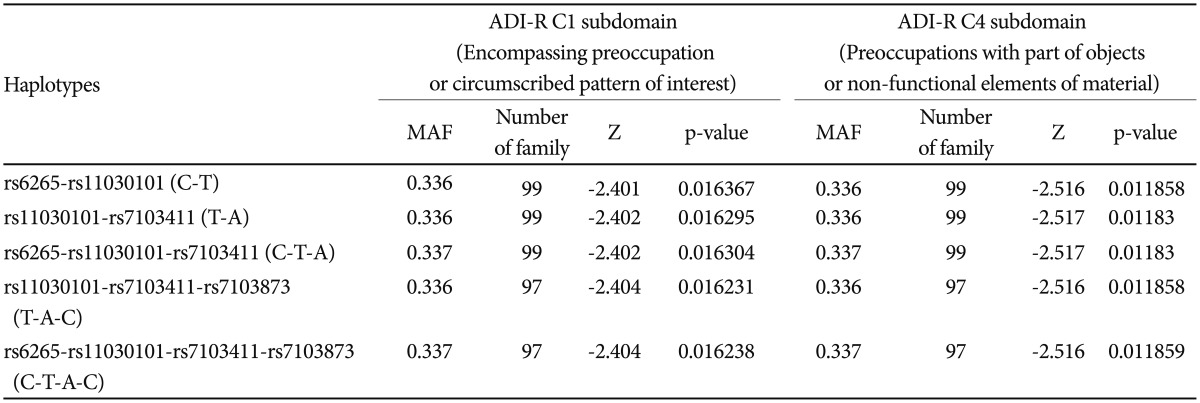
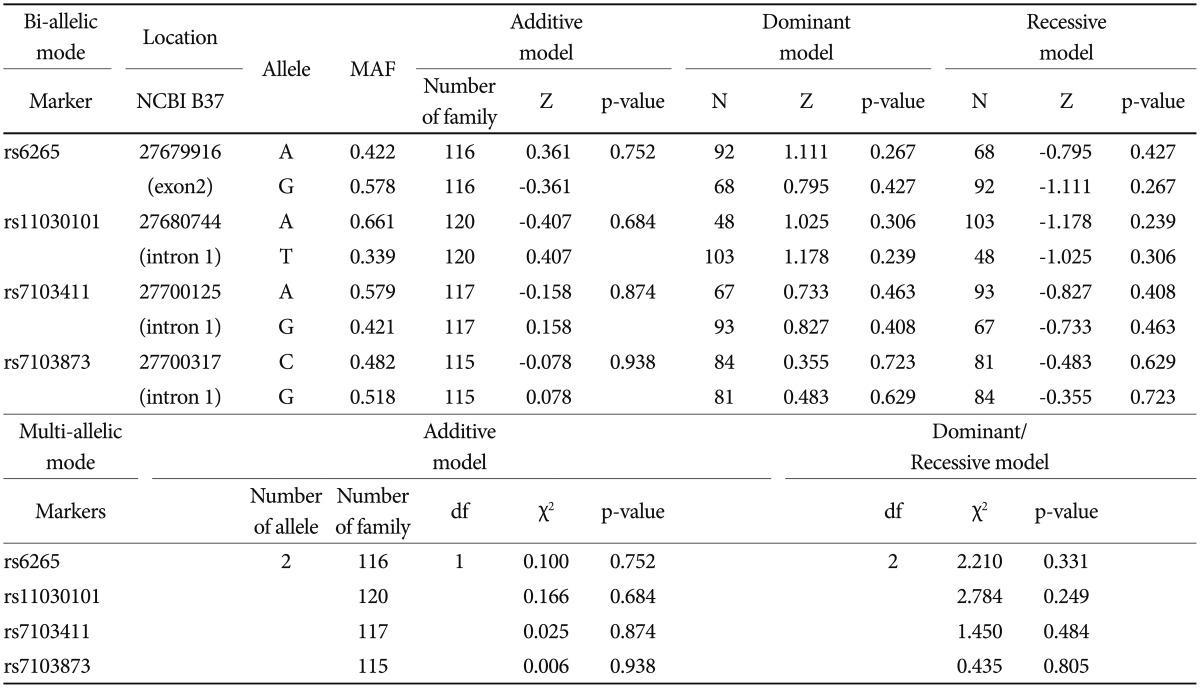
Based on FBAT analysis, no statistically significant associations were found between any polymorphisms and any of the ASD trios (Table 1). On the basis of the LD test, strong LD values (0.98<D'<0.99) were obtained for rs6265-rs11030101-rs7103411-rs7103873. In the haplotype analysis, significant associations were not observed in possible modes and models even in a sliding window approach (Table 2).
FBAT analysis results showed significant associations between the SNPs rs6265 and rs11030101 and their quantitative traits. The rs6265A allele was associated with the C1 (encompassing preoccupation or circumscribed pattern of interest) domain score within the ADI-R diagnostic algorithm in dominant/recessive models (p-value=0.019). The rs11030101 A allele was significantly associated with both C1 and C4 (preoccupations with part of objects or non-functional elements of material) domain scores within the ADI-R diagnostic algorithm in the additive model of bi- and multi-allelic modes (p-value=0.015) (Table 3). In the haplotype analysis by using quantitative traits, the sliding window method was used on 2, 3, and 4 SNP haplotype combinations. Significant associations were identified between the haplotypes and the quantitative traits from the C1 and C4 domain scores in ADI-R. Two SNP haplotypes, rs6265-rs11030101 (C-T) and rs11030101-rs7103411 (T-A) were significantly associated with the C1 (p-value=0.016) and C4 (p-value=0.012) domain scores in the additive model in bi-allelic mode. In addition, significant associations were observed between the rs6265-rs11030101-rs7103411 haplotype (C-T-A) and the rs11030101-rs7103411-rs7103873 haplotype (T-A-C) and also between the C1 (p-value=0.016) and C4 (p-value=0.012) domain scores. Similarly, the rs6265-rs11030101-rs7103411-rs7103873 haplotype (C-T-A-C) showed significant associations with the C1 and C4 domain scores (Table 4).
In this study, we showed that BDNF polymorphisms are associated with ADI-R quantitative traits in families with ASDs. To our knowledge, this is the first study showing the genetic association of BDNF gene polymorphisms and ASDs in the Korean population. The rs6265 and rs11030101 SNPs and their haplotypes were significantly associated with domains in the ADI-R category "Restricted, Repetitive, and Stereotyped Patterns of Behavior." These results not only suggest that the BDNF polymorphism is associated with autistic traits but also supports evidence of the influence of the BDNF polymorphism in the pathophysiology of ASDs.
BDNF plays an important role in neuronal systems, and the improper regulation of BDNF causes abnormal axonal and dendritic differentiation during the embryonic stages of neuronal development, as well as the formation and maturation of dendritic spines during postnatal development.19,20 The elevated serum and brain BDNF levels observed in autistic children could be attributed to a regional compensatory mechanism for the lack of development of an intrinsic component of the disease process.21 In the central nervous system, the action of BDNF is mediated by the Trk-B receptor, a member of the tyrosine kinase family of receptors.22 BDNF-TkrB signaling has a variety of endpoints, including 1) directly activating Na+, Ca2+, and K+ channels;23,24 2) enhancing glutamatergic neurotransmission by increasing the number of NMDA receptors passing through the ERK and Fyn signaling pathways;25,26 and 3) increasing the phosphorylation of the non-selective cation channel TRPC3 via PLC gamma.27 A recent study showing the screening for agents that can decrease the BDNF-TrkB pathway signaling as novel therapeutic candidates in ASDs identified a Trk partial agonist as a promising drug candidate.28 However, Scattoni et al.29 reported that BDNF and tyrosine kinase B (TrkB) protein levels in the hippocampal region were lower in the inbred BTBR T+tf/J (BTBR) strain, a putative mouse model of autism, than that in C57BL/6J mice.
Microdeletion of a 2.3 Mb region in the human chromosome 11p14.1 containing the BDNF gene has been shown to be associated with autism,30 and a whole genome scan also showed strong evidence of developmental delay.31 Recently, Han et al.11 reported that BDNF haploinsufficiency was associated with high ADI-R social interaction domain scores in 13 BDNF +/- patients with Wilms tumor, aniridia, genitourinary anomalies, mental retardation (WAGR) syndrome, a rare genetic disorder caused by heterozygous contiguous deletions of variable size in chromosome 11p13. In this study, no association was detected between BDNF SNPs and ADI-R social interaction domains, which may be attributed to the differences in sample size, ethnicity, diagnostic tools, and target polymorphisms.
The rs6265 (Val66Met) has been viewed as a representative SNP in association studies between BDNF polymorphisms and psychiatric disease, including autism.9,10 In a transmission disequilibrium test in Japanese families showing autism, the results of haplotype analysis, excluding the rs6265, by using the sliding window method showed nominally significant associations.12 However, in an association study of BDNF polymorphisms in Chinese autistic patients, the frequency of the rs6265 polymorphism was not significantly different from that of the control individuals.32 Similarly, the present study failed to identify an association between ASDs and the rs6265 polymorphism.
In this study, although only a limited number of ADI-R diagnostic algorithm domain scores were used to analyze the quantitative traits of samples, other specific traits might be present that may reveal significant associations with the SNPs in BDNF. Thus, further analysis of quantitative traits for ASDs by using biological or psychological tools is necessary. In addition, although we selected common and informative SNPs for our family-based analysis in this study, these SNPs did not exhibit the gamut of functional variants of the genetic region, and hence, future genetic analysis should be conducted using more rare variants of this gene.33
Although several associations between Korean ASD traits and BDNF gene polymorphisms were identified in this study, because of the sample size, insufficient power was observed in the analysis, which was confirmed when the results showing significant associations did not survive the Bonferroni corrections. Considering this, a large sample size with or without ethnic difference is necessary to confirm whether rs6265 and rs11030101 are risk factors for the ASD traits, namely, restricted, repetitive, and stereotyped patterns of behavior.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) (2010-0007583). This work was also supported by a research grant from Korea Healthcare Technology R&D Project (A120029) from the Ministry of Health and Welfare, Republic of Korea. Mira Park was was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2010-0012133).
References
1. Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med 1995;25:63-77. PMID: 7792363.


2. O'Roak BJ, State MW. Autism genetics: strategies, challenges, and opportunities. Autism Res 2008;1:4-17. PMID: 19360646.


3. Scherer SW, Dawson G. Risk factors for autism: translating genomic discoveries into diagnostics. Hum Genet 2011;130:123-148. PMID: 21701786.


4. Tuszynski MH, Gage FH. Neurotrophic factors and diseases of the nervous system. Ann Neurol 1994;35(Suppl):S9-S12. PMID: 8185304.


5. Huang EJ, Reichardt LF. Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci 2001;24:677-736. PMID: 11520916.



6. Miyazaki K, Narita N, Sakuta R, Miyahara T, Naruse H, Okado N, et al. Serum neurotrophin concentrations in autism and mental retardation: a pilot study. Brain Dev 2004;26:292-295. PMID: 15165668.


7. Nelson KB, Grether JK, Croen LA, Dambrosia JM, Dickens BF, Jelliffe LL, et al. Neuropeptides and neurotrophins in neonatal blood of children with autism or mental retardation. Ann Neurol 2001;49:597-606. PMID: 11357950.


8. Cardenas-Aguayo Mdel C, Kazim SF, Grundke-Iqbal I, Iqbal K. Neurogenic and neurotrophic effects of BDNF peptides in mouse hippocampal primary neuronal cell cultures. PLoS One 2013;8:e53596PMID: 23320097.



9. Gadow KD, Roohi J, DeVincent CJ, Kirsch S, Hatchwell E. Association of COMT(Val158Met) and BDNF (Val66Met) gene polymorphisms with anxiety, ADHD and tics in children with autism spectrum disorder. J Autism Dev Disord 2009;39:1542-1551. PMID: 19582565.



10. Raznahan A, Toro R, Proitsi P, Powell J, Paus T, F Bolton P, et al. A functional polymorphism of the brain derived neurotrophic factor gene and cortical anatomy in autism spectrum disorder. J Neurodev Disord 2009;1:215-223. PMID: 21547716.



11. Han JC, Thurm A, Golden Williams C, Joseph LA, Zein WM, Brooks BP, et al. Association of brain-derived neurotrophic factor (BDNF) haploinsufficiency with lower adaptive behaviour and reduced cognitive functioning in WAGR/11p13 deletion syndrome. Cortex 2013;49:2700-2710. PMID: 23517654.


12. Nishimura K, Nakamura K, Anitha A, Yamada K, Tsujii M, Iwayama Y, et al. Genetic analyses of the brain-derived neurotrophic factor (BDNF) gene in autism. Biochem Biophys Res Commun 2007;356:200-206. PMID: 17349978.


13. Yang SY, Yoo HJ, Cho IH, Park M, Kim SA. Association with tryptophan hydroxylase 2 gene polymorphisms and autism spectrum disorders in Korean families. Neurosci Res 2012;73:333-336. PMID: 22698779.


14. Yoo HJ. Korean Version of Autism Diagnostic Interview-Revised (ADIR). Seoul: Hakji-Sa; 2007.
15. Yoo HJ, Kwak Y. Korean Version of Autism Diagnostic Observation Schedule (ADOS). Seoul: Hakji-Sa; 2007.
16. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263-265. PMID: 15297300.


17. Horvath S, Wei E, Xu X, Palmer LJ, Baur M. Family-based association test method: age of onset traits and covariates. Genet Epidemiol 2001;21(Suppl 1):S403-S408. PMID: 11793708.


18. Lord C, Rutter M, Le Couter A. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord 1994;24:659-685. PMID: 7814313.


19. Fiala JC, Spacek J, Harris KM. Dendritic spine pathology: cause or consequence of neurological disorders. Brain Res Brain Res Rev 2002;39:29-54. PMID: 12086707.


20. Tyler WJ, Zhang XL, Hartman K, Winterer J, Muller W, Stanton PK, et al. BDNF increases release probability and the size of a rapidly recycling vesicle pool within rat hippocampal excitatory synapses. J Physiol 2006;574:787-803. PMID: 16709633.



21. Katoh-Semba R, Wakako R, Komori T, Shigemi H, Miyazaki N, Ito H, et al. Age-related changes in BDNF protein levels in human serum: differences between autism cases and normal controls. Int J Dev Neurosci 2007;25:367-372. PMID: 17804189.


22. Yoshii A, Constantine-Paton M. Postsynaptic BDNF-TrkB signaling in synapse maturation, plasticity, and disease. Dev Neurobiol 2010;70:304-322. PMID: 20186705.



23. Blum R, Kafitz KW, Konnerth A. Neurotrophin-evoked depolarization requires the sodium channel Na(V)1.9. Nature 2002;419:687-693. PMID: 12384689.


24. Ippolito DL, Temkin PA, Rogalski SL, Chavkin C. N-terminal tyrosine residues within the potassium channel Kir3 modulate GTPase activity of Galphai. J Biol Chem 2002;277:32692-32696. PMID: 12082117.



25. Durand GM, Kovalchuk Y, Konnerth A. Long-term potentiation and functional synapse induction in developing hippocampus. Nature 1996;381:71-75. PMID: 8609991.


26. Gomes RA, Hampton C, El-Sabeawy F, Sabo SL, McAllister AK. The dynamic distribution of TrkB receptors before, during, and after synapse formation between cortical neurons. J Neurosci 2006;26:11487-11500. PMID: 17079678.



27. Itami C, Kimura F, Kohno T, Matsuoka M, Ichikawa M, Tsumoto T, et al. Brain-derived neurotrophic factor-dependent unmasking of "silent" synapses in the developing mouse barrel cortex. Proc Natl Acad Sci U S A 2003;100:13069-13074. PMID: 14557544.



28. Tsai SJ. Is autism caused by early hyperactivity of brain-derived neurotrophic factor? Med Hypotheses 2005;65:79-82. PMID: 15893122.


29. Scattoni ML, Martire A, Cartocci G, Ferrante A, Ricceri L. Reduced social interaction, behavioural flexibility and BDNF signalling in the BTBR T+tf/J strain, a mouse model of autism. Behav Brain Res 2013;251:35-40. PMID: 23270976.


30. Autism Genome Project Consortium. Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet 2007;39:319-328. PMID: 17322880.



31. Shinawi M, Sahoo T, Maranda B, Skinner SA, Skinner C, Chinault C, et al. 11p14.1 microdeletions associated with ADHD, autism, developmental delay, and obesity. Am J Med Genet A 2011;155A:1272-1280. PMID: 21567907.


32. Cheng L, Ge Q, Sun B, Yu P, Ke X, Lu Z. Polyacrylamide gel-based microarray: a novel method applied to the association Study between the polymorphisms of BDNF gene and autism. J Biomed Nanotechnol 2009;5:542-550. PMID: 20201430.


33. Buxbaum JD. Multiple rare variants in the etiology of autism spectrum disorders. Dialogues Clin Neurosci 2009;11:35-43. PMID: 19432386.



Table┬Ā1
Family based association test and haplotype analyses with the BDNF SNPs and Korean ASD family
Table┬Ā2
Linkage disquilibrium analysis and Haplotype analyses for BDNF gene polymorphisms using sliding windows in FBAT module