The Genetic Factors Affecting Drinking Behaviors of Korean Young Adults with Variant Aldehyde Dehydrogenase 2 Genotype
Article information
Abstract
Objective
We determined whether aldehyde dehydrogenase 2 (ALDH2) activity alters the way in which drinking behaviors are affected by gene polymorphisms of other alcohol-metabolizing enzymes and serotonin-related proteins.
Methods
Through a follow-up survey with a cohort comprising 551 university freshmen over a period of 6 years, we examined the genetic factors affecting drinking behaviors. In 2000, drinking behaviors were assessed and tryptophan hydroxylase (TPH) and ALDH2 gene polymorphisms were determined. Drinking behaviors were repeated in 2006 (n=150), and the gene polymorphisms of ADH1B, ADH1C, CYP2E1, 5-HTR2A 1438A/G, and 5-HTR2A IVS2 were also determined.
Results
In 2000, the variant and wild-type ALDH2 groups exhibited little difference in terms of drinking frequency and problem drinking. Furthermore, some genotypes influenced only the variant group: ADH1B*2/*2 was associated with a lower drinking frequency, and CYP2E1 c2 allele was associated with an increased risk of problem drinking. In 2006, drinking frequency and risk of problem drinking were significantly lower in the variant group than in the wild-type group. However, the TPH AA genotype disturbed that difference, meaning that the subjects in the variant group had developed a similar level of risk of problem drinking to that in the wild-type group.
Conclusion
Korean university freshmen who were identified as a variant group drank as frequently as those in the wild-type group. For the subsequent 6 years they drank less frequently, thus decreasing the risk of problem drinking. However, that frequency drop was interrupted in those with gene polymorphisms such as ADH1B*1, CYP2E1 c2, and TPH A.
INTRODUCTION
Drinking habits tend to form in early adulthood when consuming alcohol becomes legal, and they often remain for the entire lifetime.1 Therefore, problem drinking can be effectively prevented by studying young adults' drinking behaviors and managing the identified risk factors. Problem drinking is caused by a combination of genetic, psychological, social, and cultural factors. Among these, genetic factors account for 40-60% of an individual's risk of developing problem drinking behaviors such as alcohol dependence.2
The ALDH2*2(487Lys) variant of aldehyde dehydrogenase (ALDH) genes produces a preventive effect against alcohol dependence by encoding inactive ALDH2 (ALDH2) and aggravating toxic reactions in individuals who consume alcohol.3,4 However, the impact of ALDH2 activity on drinking behavior varies among Asians. For example, when compared with other Asian American populations, Korean Americans drink more alcohol, are less sensitive to it, and are more often vulnerable to alcohol use disorders, even when they carry inactive ALDH2. These tendencies are attributable to ethnic features unrelated to alcohol-metabolizing enzymes.5,6 This makes it necessary to assess how psychosocial, psychopathological, cultural, and genetic factors influence the impact of the ALDH2 gene on alcohol drinking behavior among Koreans.7
Other alcohol-metabolizing enzymes pertaining to alcohol dependence include alcohol dehydrogenase (ADH) 1B, ADH1C, and cytochrome P450 (CYP) 2E1.8 The genes of these enzymes influence drinking behavior by accumulating and removing acetaldehyde to varying degrees depending on the activities of the enzymes expressed. ADH1B*2(*47His)9 and ADH1C*1(*350Ile)10,11 alleles express active ADH, which helps to prevent alcohol dependence, although their preventive effects are not as consistent as those of ALDH2 gene polymorphisms. The CYP2E1 c2 allele also promotes alcohol metabolism in persons with chronic drinking problems.12
A study involving the gene polymorphisms of tryptophan hydroxylase (TPH), an enzyme that limits the oxidation rate of tryptophan into serotonin, found that the A allele can be associated with early-onset alcohol dependence.13 Therefore, problem drinking may develop early in life under the influence of serotonin-related gene polymorphisms, including TPH. Some psychopathological factors often influence this type of alcoholism, including impulsivity and criminal tendencies.
In a previous study conducted in 2000 on 551 university freshmen, we identified a relationship between drinking behaviors and ALDH2 and TPH genotypes.14 We also examined how changes in their drinking behavior in the subsequent 6 years were related to ALDH2 activity15 and psychosocial stress.16 The present study was designed as a follow-up to that study, to identify whether drinking behavior is affected differently in subjects with different types of ALDH2 and other genetic polymorphisms of alcohol-metabolizing enzymes such as ADH1B, ADH1C, and CYP2E1, and those of serotonin-related proteins such as TPH, 5-HTR2A 1438A/G, and 5-HTR2A IVS. By identifying whether there exist other genetic factors that encourage drinkers to continue consuming alcohol despite the adverse reactions caused by variant ALDH2 genes, this study sought to understand drinking behaviors that are unique to Koreans, and the risks presented by such behaviors in order to provide information that can be used to devise countermeasures against problem drinking.
METHODS
Subjects
The cohort in this study was the same as that in our previous study (n=551 persons; 308 males and 243 females).14 As of March 2006, when the follow-up study was initiated, 293 of these were still at school and the remaining 241 were not. This follow-up study was therefore conducted on 150 (102 males and 48 females) of the original subjects.
Procedures
Before the study was initiated, approval was obtained from the Bioethics Committee in Chungbuk National University College of Medicine. To obtain the consent of the subjects of the 2000 study for this additional research, those still attending school were contacted with the assistance of the university's administrative office, informed of the purpose of the study, and asked to complete a survey. To obtain the consent of the graduates, the survey was mailed to their most recent home addresses, which were obtained with the help of the administrative office and the alumni association. The study was carried out between March and September 2006.
Measurements
Retaining the same basic categories used in the previous study's survey questions, the 2006 survey included questions on the subjects' demographic features, drinking severity, and drinking behavior. To evaluate drinking severity and behavior, a questionnaire developed in 1998 by the Korea Institute for Health and Social Affairs, entitled the "Korea National Health and Nutrition Examination Survey,"17 was revised by incorporating parts of the CAGE questionnaire18 and the Korean version of the Alcohol Use Disorder Identification Test (AUDIT-K).19 As in the previous study, the drinking severity index, which quantifies drinking severity on a scale of 1 to 30, is calculated by multiplying a frequency score (from 1 to 6 points) by an amount score (1 to 5 points).
Genotyping
The results of the previous study14 on ALDH2 and TPH genotypes were utilized once again in this study. Further information was obtained by performing additional experiments on the subjects' DNA, which had been obtained in 2000 and preserved in a frozen state after obtaining the subjects' consent. In 2006, genotyping was carried out using an ABI SNaPshot Multiplex Kit (ABI, Foster City, CA, USA) according to the manufacturer's recommendations. Following amplification, removal of primers and dNTP, and addition of extension primers to six SNP sites, SNaPshot products were run on an ABI 3100 system and analyzed using GeneScan 3.1 software (ABI). The details of the genotyping method have been described previously.20
Data analysis
Based on their ALDH2 genotypes, the subjects were divided into the wild-type ALDH2 gene group (ALDH2*1/*1, wild-type group) and the variant ALDH2 gene group (ALDH2*1/*2, *2/*2, variant group). Chi-square tests were performed to confirm whether the genotypes of proteins such as ADH1B, ADH1C, CYP2E1, TPH A218C, TPH2 1463G/A, 5-HTR2A 1438A/G, and 5-HTR2A IVS2 are relevant to drinking frequency and amount, and whether the probability of being classified by CAGE and AUDIT-K as being at risk of developing a drinking problem (henceforth referred to as simply "at risk") varied between the wild-type and variant groups. When differences among groups showed a trend (p≤0.10), Fisher's exact tests were conducted after merging multiple cells of a 2×2 table. In the process of merging, subjects who achieved CAGE21 scores of 2 points or higher and AUDIT-K19 scores of 12 points or higher were identified as being at risk. The median values of drinking frequency and amount scores were utilized to categorize subjects as follows: those who drink once or less in a week and those who drink twice or more (for the frequency); and those who drink one bottle of soju or less at a time and those who drink more than one bottle at a time (for the amount). Among Koreans, soju is a popular liquor containing 20-25% alcohol by volume. One bottle of soju contains nearly seven units of alcohol.
Based on the obtained results, we performed logistic regression analysis: covariants were the genes confirmed as being associated with ALDH; dependent variables were the groups identified based on the risk of developing problem drinking (high-risk and low-risk groups), by frequency (those who drink once or less a week, and twice or more), and by drinking amount (those who drink one bottle or less at a time, and more than a bottle). The data-analysis process is shown in Figure 1.
RESULTS
Gene distribution
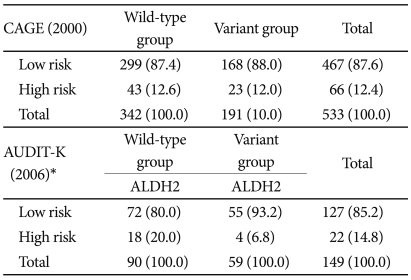
The distributions of ADH1B, ADH1C, CYP2E1, TPH A218C, TPH2 1463 A/G, 5HTR2A 1438A/G, and 5-HTR2A IVS2 among the subjects and their Hardy-Weinberg equilibria are given in Table 1. The study performed in 2000 analyzed the responses from 533 out of the 534 subjects whose ALDH2 genotypes were determined; the single excluded subject exhibited insufficient responses. Among these 533 subjects, 342 (64.2%) were wild-type and 191 (35.8%) were variant. Sixty-six subjects (12.4%) were identified by CAGE as being at risk, and their ALDH2 genotypes did not affect that identification. Among the 150 subjects included in the 2006 study, 149 were analyzed and identified as either wild-type (n=90, 60.4%) or variant (n=59, 39.6%). AUDIT-K identified 22 subjects (14.8%) as being at risk; the ALDH2 genotype was a significant factor in that identification (Table 2).
Genetic factors affecting drinking frequency and amount
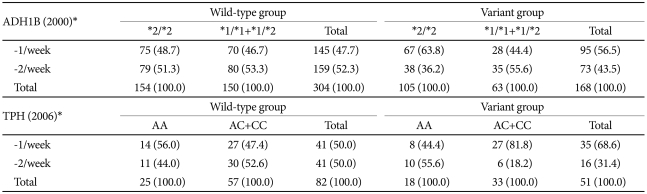
In each ALDH2 genotype group we searched for genetic factors that varied with the drinking frequency. In 2000, the ADH1B genotype did not influence the drinking frequency of the wild-type group, but the frequency in the variant group was lower when the ADH1B*2/*2 allele was present. In the 2006 study, the TPH genotype produced no frequency differences in the wild-type ALDH2 group, whereas the TPH C allele reduced the drinking frequency in the variant group (Table 3). With the exception of ALDH2, none of the studied genes affected the drinking amount (i.e., no effect was observed in either the wild-type or variant group).
Genetic factors affecting identification of at-risk subjects
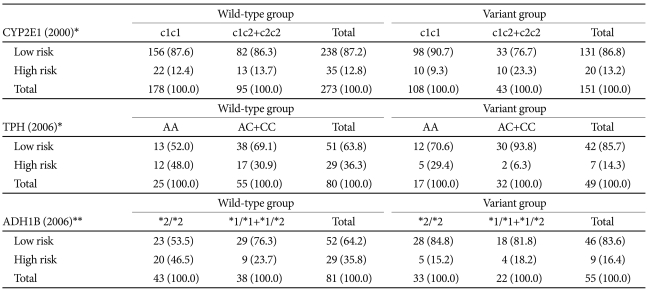
The genetic factors affecting the probability of being identified as being at risk (i.e., subjects scoring 2 points or higher in CAGE and 12 points or higher in AUDIT-K) were investigated in both the ALDH2 genotype groups. In the 2000 study, the CYP2E1 genotype had a differential impact on the wild-type and variant ALDH2 groups, whereby no impact was observed in the wild-type group, whereas in the variant group the probability of being identified as being at risk increased when the subjects carried the CYP2E1 c2 allele. In 2006, TPH and ADH1B were found to be significant. TPH did not affect the probability of wild-type subjects being classified as at risk, but it considerably reduced that probability among subjects with the C allele in the variant group. The ADH1B genotype was benign in the variant group, but increase the probability of being identified as being at risk among those with ADH1B*2/*2 in the wild-type group (Table 4).
Logistic regression analysis
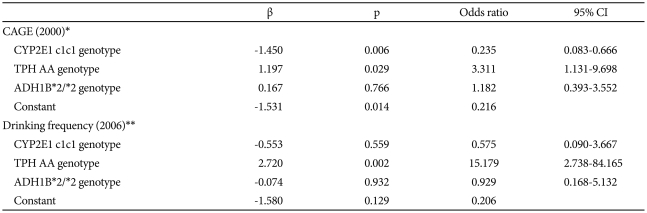
Logistic regression analysis was carried out using the CYP2E1 c1c1, TPH AA, and ADH1B*2/*2 genotypes as covariants, all of which were assumed to influence drinking behavior based on the aforementioned observations. In 2000, the CYP2E1 c1c1 and TPH AA genotypes were found to be significant genetic factors influencing the CAGE-based identification of being at risk in the variant group. It was found that the TPH AA genotype was still a factor in 2006, increasing the drinking frequency in the variant group. The CYP2E1 c1c1 genotype reduced the probability of being identified as at risk by 0.24 times in 2000, while the TPH AA genotype was shown to increase that probability in 2000 and to increase the drinking frequency in 2006 by about 3.3 and 15.2 times, respectively (Table 5).
DISCUSSION
The 2000 study revealed that the ALDH2 genotype does not affect an individual's probability of being identified by CAGE as either being at risk or being a frequent drinker, although relevant data for the latter were not reported. However, the ALDH2 genotype was found to influence the drinking amount. For example, 52.6% of the subjects in the wild-type group drank more than one bottle of alcohol at a time, while only 32.2% of the variant group did so.14 University freshmen with different ALDH2 genotypes showed no difference in drinking frequency, since they drank mainly in response to social pressure. Moreover, once they started drinking, these subjects continued until they reached a similar level of drunkenness, which is contingent on ALDH2 activity. Therefore, the subjects presented similar physiological responses and drinking frequencies, regardless of their ALDH2 genotype, albeit with some differences caused by ALDH2 activity. As a result, subjects with active and those with inactive ALDH2 responded to the CAGE questionnaire similarly in terms of whether they felt the need to cut down on drinking or whether they ever felt guilty about drinking. However, in the 6 years since the 2000 study, there have been some changes to these observations, since the subjects appear to have modified their drinking frequency depending on their ALDH2 activity, to arrive at different levels of risk of developing a drinking problem.
In 2000, it was found that the impact of ADH1B on drinking frequency differed depending on ALDH2 genotype. While ADH1B did not reduce drinking frequency among the wild-type subjects, ADH1B*2/*2 decreased the drinking frequency among those in the variant group. Meanwhile, subjects with ADH1B*1/*1 and ADH1B*1/*2 in the variant group drank as frequently as their wild-type counterparts. The variant ALDH2 gene alone cannot reduce the drinking frequency among freshmen, since this is determined by social pressures; drinking frequency is reduced when the gene works together with the ADH1B*2/*2 genotype. The ADH1B*2 allele expresses the β2 subunit, whose turnover rate is 70-80 times higher than that of the β1 subunit expressed by the ADH1B*1 allele.22 Therefore, unlike ADH1B*1/*1, the ADH1B*1/*2 and ADH1B*2/*2 genotypes exert preventive effects against alcohol-use disorder. However, since the study included very few subjects with the ADH1B*1/*1 genotype, the gap in drinking frequency observed was attributable to the differences between ADH1B*1/*2 and ADH1B*2/*2. The differences in ADH1B activity expressed by the two genotypes were not distinctive enough to produce a gap in alcohol-dependence risk.23 Yet, even this subtle difference produced a significant impact in the variant group (but not the wild-type group) in early adulthood.
The CYP2E1 gene did not affect the risks of problem drinking identified by the CAGE in the wild-type group, while the CYP2E1 c2 allele did increase the risk in the variant group. CYP2E1 influences alcohol metabolism in chronic drinkers, with the c2 allele increasing it.12 Therefore, when CYP2E1 activity increases in the members of the variant group, they overcome their weak alcohol metabolism and continue problem drinking. This results in the generation of high levels of toxic substances such as free radicals. Therefore, those with both the CYP2E1 c2 allele and the variant ALDH2 gene who continue problem drinking are especially susceptible to the physical damage caused by alcohol consumption.
CYP2E1 did not produce any differences in the wild-type group, whose members promptly metabolize toxic substances. However, among the variant group, even subtle physiological responses caused by CYP2E1 or ADH1B genotypes produced a difference in drinking behavior. Sun et al.24 reported that the risk of excessive drinking increases in Japanese males aged 23-64 years when they carry both the ALDH2*1 and CYP2E1 c2 alleles. However, the risk of regular excessive drinking increased among young Koreans when they carried the ALDH2*2 geno type and the CYP2E1 c2 allele. Such a difference, which appears to be linked to race, age, and research methods, requires additional research.
In the 2006 study, the impact of TPH genotype on drinking frequency differed with the ALDH2 activity. In the wild-type group, nearly 50% of subjects drank two or more times in a week, regardless of their TPH genotype. Meanwhile, the proportion of subjects who drank at the same frequency fell to 18.2% among those with the TPH C allele in the variant group, although the variant and wild-type groups had similar proportions of members carrying TPH AA.
Furthermore, the TPH genotype was found to have an impact on problem drinking. Those in the wild-type group presented a 36.3% risk of being identified by AUDIT-K as being at high risk, regardless of their TPH genotype. In contrast, the risk was lower in the variant group, dropping to as low as 6.3% among those with the TPH C allele, but to only 29.4% among those carrying the TPH A allele. These results indicate that the TPH AA allele restrains the process by which inactive ALDH2 inflicts the adverse reaction that reduces problem drinking.
When they began drinking alcohol in 2000, those in the variant group drank as frequently as their counterparts in the wild-type group. However, over time, they reduced their drinking frequency, and by 2006 many had ceased to be classified as being at risk. As noted above, the TPH AA genotype interferes with the reduction in drinking frequency, which is why people with the genotype of the variant group are at a higher risk of developing a drinking problem.
A study that examined Korean patients with alcohol dependence found that the TPH A allele is associated with early onset of alcohol dependence in people aged 25 years and older.13 This study again confirmed the gene's effect, since all of the subjects became older than 25 years during the subsequent 6-year period. This pathogenesis could be attributed to serotonin-related psychosocial factors such as impulsivity, aggression, and depression.
The results of the logistic regression analysis are similar to those of the aforementioned study.13 The 2000 study devised a valid model of factors that help to identify those with a risk of problem drinking only in the variant group. According to the model, the CYP2E1 c1c1 genotype decreases the probability of being classified as a problem-drinker. When CYP2E1 activity that bypasses ADH and ALDH metabolism is reduced, genetically weak alcohol metabolism resulting from the variant group's inactive ADLH2 combines with an intensified adverse reaction after drinking to reduce the probability of developing a drinking problem. In contrast, TPH AA increases the probability of being classified as being at risk by helping to overcome weak alcohol metabolism through psychosocial mechanisms, and thus of continuing drinking.
The 2006 study established a valid model of factors identifying frequent drinkers that applies only to the variant group. According to the model, the TPH AA genotype increases the probability of being included in a frequent-drinker's group. Subjects with inactive ALDH2 suffered an adverse reaction after drinking, which resulted in most of them reducing their drinking frequency over the 6-year study period. However, those in the variant group with the TPH AA genotype were able to resist drinking-related distress and maintain their drinking frequency, leading them to be identified as being at risk.
This study was subject to several limitations. The study tracked only a small number of demographically homogeneous people who were the subjects of the 2000 study. In fact, it was easier to track down subjects who remained in school than to locate the entire original cohort that had graduated and become scattered throughout the country. For that reason, the 2006 study included twice as many student subjects as graduates. It also included more male than female subjects, because male students returned to school after completing their military service, while the women-who do not serve in the South Korean military-tended to graduate earlier than their male counterparts. Thus, the results of the 2006 study might apply only to Korean men in their late twenties. Going forward, a more comprehensive and systematic approach is needed to complement these weaknesses.
Furthermore, different tools were used to identify the at-risk group in the 2000 and 2006 studies: CAGE in 2000 and AUDIT-K in 2006. CAGE was utilized in 2000 because AUDIT-K was not widely used at that time. Since CAGE and AUDIT-K might define aspects of problem drinking differently, caution is required when directly comparing and interpreting the results of the two studies. However, since both are established tests used in alcoholism screening, it would be ideal to utilize both in future follow-up studies and to compare the results.
Despite its limitations, this study is meaningful in that it tracked the drinking behavior of Koreans starting from the age when it became legal for them to drink alcohol, and comprehensively identified the factors affecting their drinking behavior. It can be concluded that Korean university freshmen with the variant ALDH2 genotype drink just as frequently as those with the wild-type ALDH2 genotype. Although the members of variant group on average drink less than the wild-type group, both experience similar physical and psychological effects after drinking due to the weaker alcohol-metabolizing capability of the former. Therefore, unlike those in older age groups with the variant ALDH2 genotype who drink less and present a lower risk of developing problem drinking than those with the wild-type gene, young adults with the variant genotype do not appear to experience a reduced risk. However, by their late twenties their risk falls to a lower level than that of the wild-type group due to less-frequent drinking. However, if other metabolizing enzymes such as ADH1B and CYP2E1 compensate for the weaker alcohol metabolizing capability, or if TPH and psychosocial factors work together to interrupt the drop in drinking frequency, the risk of problem drinking does not decrease. Further studies are needed in order to elucidate the psychosocial features and alcohol metabolism of those at risk of problem drinking.