CDH13 and HCRTR2 May Be Associated with Hypersomnia Symptom of Bipolar Depression: A Genome-Wide Functional Enrichment Pathway Analysis
Article information
Abstract
Although bipolar disorder is highly heritable, the identification of specific genetic variations is limited because of the complex traits underlying the disorder. We performed a genome-wide association study of bipolar disorder using a subphenotype that shows hypersomnia symptom during a major depressive episode. We investigated a total of 2,191 cases, 1,434 controls, and 703,012 single nucleotide polymorphisms (SNPs) in the merged samples obtained from the Translational Genomics Institute and the Genetic Association Information Network. The gene emerging as the most significant by statistical analysis was rs1553441 (odds ratio=0.4093; p=1.20×10-5; Permuted p=6.0×10-6). However, the 5×0-8 threshold for statistical significance required in a genome-wide association study was not achieved. The functional enrichment pathway analysis showed significant enrichments in the adhesion, development-related, synaptic transmission-related, and cell recognition-related pathways. For further evaluation, each gene of the enriched pathways was reviewed and matched with genes that were suggested to be associated with psychiatric disorders by previous genetic studies. We found that the cadherin 13 and hypocretin (orexin) receptor 2 genes may be involved in the hypersomnia symptom during a major depressive episode of bipolar disorder.
INTRODUCTION
Bipolar disorder (BD) patients suffer from both a low physical quality and low mental quality of life, even during an euthymic period.1 BD is known to be highly heritable; however, the identification of specific genetic variations resulted in limited findings.23 One reason for this low success rate in identifying genetic variations relies in the nature and complex traits of BD.4 As a consequence, dividing BD into subgroups according to clinical subphenotypes is a reliable alternative approach for future genetic studies in BD.5
Several studies suggested that dysregulation of the circadian rhythm is associated with the pathophysiology of BD.67 In general, BD patients have common specific circadian rhythm-related clinical manifestations such as: diurnal mood variation, periodicity of manic-depressive episodes, activity changes, alteration of the secretion of hormones and other endogenous substances, and sleep disturbances.8910 Sleep disturbances may have a negative effect on emotional regulation.11 According to these results, it can be assumed that circadian rhythm dysregulation is very closely associated with BD.12
We hypothesized that a subphenotype group that shows hypersomnia symptom during a major depressive episode of bipolar disorder may represent a genetically distinct subtype of BD. We tested this hypothesis using a genome-wide association (GWA) analysis between subjects that showed hypersomnia symptom during a major depressive episode of bipolar disorder (hypersomnia symptom of bipolar depression, HBD) and subjects that did not show hypersomnia symptom during a major depressive episode of bipolar disorder (non-hypersomnia symptom of bipolar depression, NHBD). The subject samples were of European ancestry and were genotyped as part of the Genetic Association Information Network (GAIN) by the Bipolar Genome Study (BiGS).
METHODS
Subject ascertainment
Prior to genotyping, which was part of the BiGS, the unrelated bipolar I disorder subjects of European ancestry were selected from those collected by the National Institute of Mental Health Genetics Initiative for Bipolar Disorder. All subjects provided written informed consent in accordance to protocols from local institutional review boards. The subjects were interviewed using protocols from the Diagnostic Interview for Genetic Studies (DIGS).13 Information was obtained from family informants and medical records. The information was reviewed along with the interview by a panel of experienced clinicians to obtain a final best-estimate diagnosis. Control subjects were selected after they were certified through a National Institute of Mental Health-supported contract between Dr. Pablo Gejman and Knowledge Networks, Inc.14 All subjects donated a blood sample and were given medical questionnaires. The selected controls were matched for gender and ethnicity with the BD cases. Control subjects who had a history of BD, psychosis, or recurrent major depression were excluded from our study. The subject samples as part of the GAIN were obtained by Dr. Kelsoe who is a member of the BiGS.
Genotyping and cleaning
The first set of samples was genotyped at the Broad Institute, as part of GAIN, using the Affymetrix SNP Array 6.0 (Affymetrix; Santa Clara, CA, US) 1M SNP array. We obtained a total of 1,001 BD cases, 1,033 controls, and 724,067 single nucleotide polymorphisms (SNPs). These were available for analysis following an extensive quality control (QC) process.15 The QC process eliminated all individuals with >10% missing data, SNPs with poor allele clustering, duplicate errors, minor allele frequencies <0.05, and significant deviation from Hardy-Weinberg equilibrium at p<10-6. The second set of samples was genotyped similarly to the first set of samples at the Translational Genomics Institute (TGEN) and underwent a comparable QC. From this set of samples we obtained 1,190 BD cases, 401 controls, and 728,187 SNPs available for analysis. An additional round of QC was performed on the merged samples from GAIN and TGEN. This merge resulted in a set of 703,012 SNPs that passed the imposed QC process.
Phenotypes
As part of the DIGS interview, bipolar I disorder subjects were queried as to whether they had a hypersomnia symptom during major depressive episodes. According to the answer, subjects were categorized either into the HBD or the NHBD group. Those who answered 'Unknown' to the question about hypersomnia symptom were categorized as the missing group. After filtering, there were 263 BD subjects in the HBD group and 112 subjects in the NHBD group.
Association analyses
To assess genetic factors contributing to HBD, we performed a genome-wide case-only analysis of HBD versus NHBD. This association analysis was performed using a logistic regression using PLINK16 with a covariance adjustment for sex and age. Adaptive permutations were performed to find the empirical significance of the results using PLINK.16
SNP imputation
Missing SNPs were imputed using the IMPUTE2 tool17 and the CEU panel of HapMap 3+1,000 Genomes Pilot haplotypes as a reference. The imputed SNPs were used for the functional enrichment pathway analysis.
Functional enrichment pathway analysis
Gene ontology analysis was performed on the gene sets harboring the identified SNPs (p<0.005) evaluated using the DAVID software.1819 The enriched gene functions were identified from the HBD versus NHBD analysis using an enrichment score. To go further in our investigation, we made a list of genes of the enriched pathways and compared them with various genes that were suggested to be related to psychiatric disorders by previous genetic studies. We searched the available databases in PubMed (http://www.ncbi.nlm.nih.gov/pubmed/) and in the National Human Genome Research Institute (http://www.genome.gov/) for genes of enriched pathways. Some of the search terms included: the name of each gene of enriched pathways and "psychiatric disorder or depression or bipolar disorder or schizophrenia or circadian rhythm". After the review and matching, we selected genes from those of enriched pathways that were described as susceptible genes in psychiatric disorders by previous genetic studies.
RESULTS
Eighty associated SNPs with p<10-4 were identified in this present genome-wide case-only analysis. We found that rs1553441 is the gene that appeared to be the most significant after statistical analysis (odds ratio=0.4093, p value of 1.20×10-5, Permuted p=6.0×10-6) (Supplementary Figure 1 in the online-only Data Supplement). This SNP is located within a region of the gene encoding the phospholipase D family, member 5 (PLD5) on chromosome 1q43. However, the most significant gene of this study did not reach the p value of 5×10-8, which is the threshold for statistical significance in a GWA.16
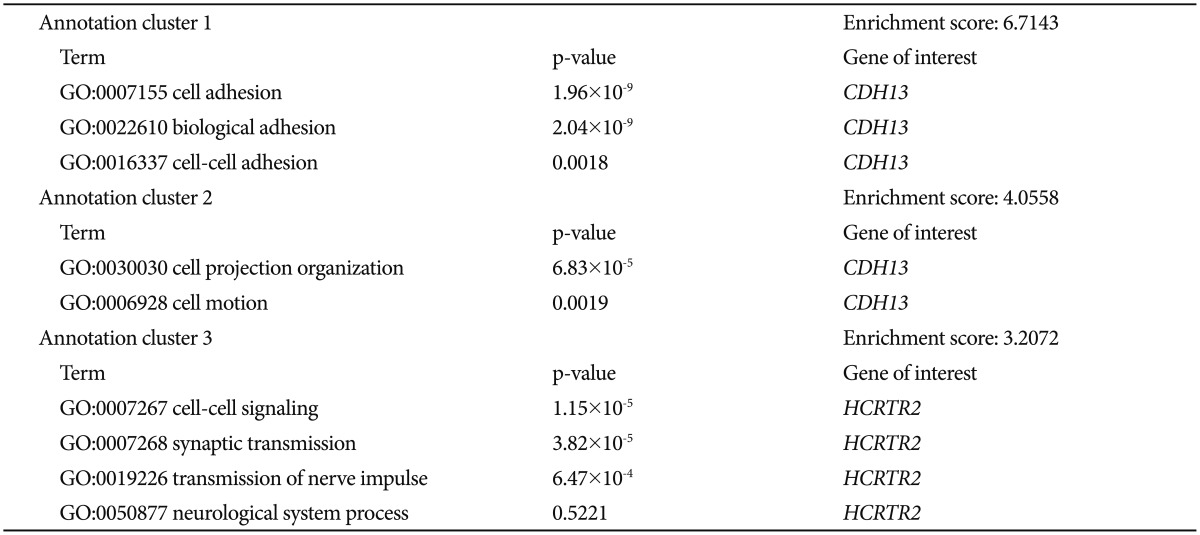
To determine whether the associated SNPs showed enrichment in certain functional pathways, we performed a genome-wide functional enrichment pathway analysis using the imputed data. Gene ontology functions were identified for all genes that contained SNPs associated with a significance level of p<0.005. The functional enrichment analysis of HBD versus NHBD showed a significant enrichment of the adhesion, development-related, synaptic transmission-related, and cell recognition-related pathways (Supplementary Table 1 in the online-only Data Supplement). We can speculate that the altered functions of these pathways may contribute to the development of hypersomnia symptom during a major depressive episode of BP.
We aimed to investigate the genes of the enriched pathways in more detail. At first, we reviewed the results of previous genetic studies of psychiatric disorders found in the PubMed database (http://www.ncbi.nlm.nih.gov/pubmed/) and the database from the National Human Genome Research Institute (http://www.genome.gov/). To search the databases we used search terms such as: the name of each gene of enriched pathways and "psychiatric disorder or major depressive disorder or bipolar disorder or schizophrenia or circadian rhythm". In accordance with previously reported genetic findings, we investigated the significant genes of those identified from categories that showed significant enrichment by comparison to the results of previous genetic studies. We selected the genes of enriched pathways that matched those that have been suggested in the literature to be related to psychiatric disorders (Table 1). We ultimately found two matching genes: cadherin 13 (CDH13) and hypocretin (orexin) receptor 2 (HCRTR2). CDH13 is located in 16q24.2 and encodes a member of the cadherin superfamily. The encoded protein acts as a negative regulator of axon growth during neural differentiation.2021 In the present pathway analysis, CDH13 was identified as a member of genes thought to be involved in cell adhesion, biological adhesion, cell-cell adhesion, cell projection organization, and cell motion. HCRTR2 is located in 6p12 and encodes a G protein-coupled receptor involved in regulation of feeding and sleep behavior.22 In the present pathway analysis, HCRTR2 was identified as a member of genes thought to be involved in cell-cell signaling, synaptic transmission, transmission of nerve impulse, and neurological system process.
DISCUSSION
We were not able to identify significant genes in a GWA study that evaluated HBD versus NHBD. As a consequence, we focused on the pathways identified in genome-wide functional enrichment pathway analysis that showed significant enriched function. Specifically, we evaluated the gene lists of these enriched pathways, and compared them with genes that were previously suggested in the literature to be related to psychiatric disorders. This procedure was found to be helpful in finding genes that are possibly related to HBD, although this is an unfamiliar method and the results do not always provide an explanation on how genes are related.
CDH13 is highly expressed in various brain regions such as the cerebral cortex, medulla, thalamus, and midbrain.20 It has been suggested that CDH13 is associated with various drug abuse-related phenotypes,232425262728 especially with respect to comorbid depression and alcohol dependence.29 Disorders related to substance use, particularly alcohol dependence, show high comorbidity with BD.30 In addition, the presence of substance abuse disorders seriously affects the course and prognosis of BD.31 CDH13 expression also showed an association with adiponectin levels3233 and depression.34 Previous studies investigated adult depressed patients that showed decreased circulating levels of adiponectin.353637 One explanation for this finding could be that adiponectin is related to the Hypothalamic-Pituitary-Adrenal (HPA) axis. Several studies suggested a correlation between adiponectin and stress and development of various psychiatric disorders.38 Furthermore, TNF-α, IL-10, and IL-6, previously shown to be regulated by adiponectin, also showed a correlation with BD.3940 TNF-α is suggested to produce an increased slow-wave sleep and a symptom of sleepiness.41 Orexin, also known as hypocretin-A and B, neuromodulatory peptides secreted from orexin neurons, signal through orexin receptor 1 and orexin receptor 2 (HCRTR1 and HCRTR2), which are G protein-coupled receptors.42 Orexin-containing neurons project out to various monoaminergic nuclei of the brain, including the locus coeruleus (norepinephrine), the raphe nuclei (5-hydroxytryptamine, 5-HT), and the ventral tegmental area (dopamine). Therefore, the orexin-containing neurons can modify the monoaminergic neurons.43 Furthermore, the presence of a functional positive feedback loop between the orexin and monoaminergic neurons has been suggested.44 Orexin is implicated in diverse functions such as feeding, drinking, the sleep-wake cycle, hormone secretion, autonomic function, drug addiction, and reward. To date, most research has focused on the role of orexin in depression compared to other mood disorders, and several studies suggested that orexin plays a significant role in depression.45 In particular, several studies have reported a role for orexin in sleep disturbance,4647 reward system,48 feeding behavior,4950 hippocampal neuronal plasticity,515253 and monoamines.5455 It is important to note that BP is an important portion of depressive symptoms. Moreover, atypical depressive symptoms, characterized by hypersomnia, hyperphagia (or weight gain), and leaden paralysis, are more common in patients with BP than unipolar depression.56 We speculate that representative symptoms of atypical depression, such as hypersomnia and hyperphagia, may be closely related to orexin's function. Moreover, a balanced effect of orexin's action on either the HCRTR1 or the HCRTR2 receptor is important in achieving an anti-depressant- or pro-depressant-like effect.57 Taken together, these previous findings suggest that hypersomnia symptoms during a major depressive episode of bipolar disorder are related to the functions of CDH13 and HCRTR2.
We have explored the genetic association of BD using the HBD subphenotype to focus on a more homogenous group of subjects that present similar clinical courses. Although the present GWA of HBD versus NHBD did not produce significant results, the functional enrichment pathway analysis identified significant enrichments of the adhesion, development-related, synaptic transmission-related, and cell recognition-related pathways. In addition, we found that CDH13 and HCRTR2 show a potential correlation with HBD and we used a matching process based on the results of previous genetic studies performed on psychiatric disorders. To date, only several studies have been reported about this correlation. In the future, more studies focusing on the correlation between these pathways or several genes and HBD are needed to validate the results reported in the present study.
Acknowledgments
Bipolar Genome Study Co-authors: John R. Kelsoe, Tiffany A. Greenwood, Caroline M. Nievergelt, Rebecca McKinney, Paul D. Shilling, Erin N. Smith - University of California, San Diego, CA, USA; Nicholas J. Schork, Cinnamon S. Bloss - Scripps Translational Science Institute, La Jolla, CA, USA; John I. Nurnberger, Jr., Howard J. Edenberg, Tatiana Foroud, Daniel L. Koller - Indiana University, Indianapolis, IN, USA; Elliot S. Gershon, Chunyu Liu, Judith A. Badner - University of Chicago, Chicago, IL, USA; William A. Scheftner - Rush University Medical Center, Chicago, IL, USA; William B. Lawson, Evaristus A. Nwulia, Maria Hipolito - Howard University, Washington, D.C., USA; William Coryell - University of Iowa, Iowa City, IA, USA; John Rice - Washington University, St. Louis, MO, USA; William Byerley - University of California, San Francisco, CA, USA; Francis J. McMahon, Thomas G. Schulze - National Institute of Mental Health Intramural Research Program, Bethesda, MD, USA; Wade H. Berrettini - University of Pennsylvania, Philadelphia, PA, USA; James B. Potash, Peter P. Zandi, Pamela B. Mahon - Johns Hopkins School of Medicine, Baltimore, MD, USA; Melvin G. McInnis, Sebastian Zöllner, Peng Zhang - University of Michigan, Ann Arbor, MI, USA; David W. Craig, Szabolics Szelinger - The Translational Genomics Research Institute, Phoenix, AZ, USA; Thomas B. Barrett - Portland Veterans Affairs Medical Center, Portland, OR, USA
This work was supported by grants from the National Institute of Mental Health (NIMH) and National Human Genome Research Institute (NHGRI) to JRK (MH078151, MH081804, and MH059567supplement), and by the Genetic Association Information Network (GAIN). HJL was supported by the Korea Health 21 R&D Project funded by the Ministry of Health & Welfare, Republic of Korea (HI11C1901) and the Future Environmental R&D grant funded by the Korea Environmental Industry and Technology Institute (No. RE201206020).
References
Supplementary Materials
The online-only Data Supplement is available with this article at http://dx.doi.org/10.4306/pi.2015.12.3.402.
Supplementary Figure 1
Genome-wide association results for hypersomnia bipolar depression. The Manhattan plot shows the results of susceptible loci relevant to hypersomnia bipolar depression. The chromosomal position is shown along the X-axis, whereas the -log (p value) for each single nucleotide polymorphism is shown along the Y-axis. The horizontal line indicates the p<10-4 significance threshold. The arrows in the figure indicate the position of possible relevant genes above the set threshold.
Supplementary Table 1
Enrichment findings from the genome-wide functional enrichment pathway analysis of hypersomnia bipolar depression