Target Discovery Using Deep Learning-Based Molecular Docking and Predicted Protein Structures With AlphaFold for Novel Antipsychotics
Article information
Abstract
Objective
New drugs are needed to treat antipsychotic-resistant schizophrenia, especially those with clozapine-resistant schizophrenia. Atypical antipsychotics have predominantly 5-HT2A and dopaminergic antagonism, but also require investigation of other receptors.
Methods
In this study, the binding affinities between clozapine, olanzapine, and quetiapine with neuropharmacological, immunological, and metabolic receptors were measured using GNINA (Deep Learning Based Molecular Docking) and AlphaFold (Predicted Protein Structures).
Results
Through this study, it was determined that these antipsychotics showed high binding affinity to a variety of receptors, such as CB2, 5-HT1BR, NPYR4, and CCR5. Cyclosporin A and everolimus which show high affinities with those receptors could be used for the development of new antipsychotic drugs based on these drugs.
Conclusion
In the future, the method used in this study will be applied to the development of new antipsychotic drugs, including drug repositioning, and to the discovery of the pathophysiology of schizophrenia.
INTRODUCTION
Schizophrenia is a chronic mental illness accompanied by psychotic symptoms such as hallucination and delusion, negative symptoms such as avolition and affective blunting, and cognitive impairment. This condition is the eleventh leading cause of years lived with disability in 2013 [1,2]. Drugs for psychiatric disorders, including schizophrenia, mainly act on monoamine receptors, and have an overall therapeutic effect of approximately 50%. Intractable cases occur in about 20% of patients [3,4]. Factors that include low drug adherence, as well as psychiatric and medical comorbidities, contribute to drug treatment resistance. However, antipsychotics resistance appears even in schizophrenia patients with high adherence to treatment and no comorbidities, which results in unmet medical demand [5].
Clozapine is an antipsychotic drug that has the best effect on auditory hallucinations and delusions among antipsychotics, and has fewer extrapyramidal side effects such as the Parkinsonian-like behavior induced by antipsychotic drugs [6]. Clozapine was shown to be effective in about 70% of patients who were resistant to antipsychotic drugs other than clozapine. However, in a previous study, about 50% of schizophrenia symptoms persisted for more than 12 years among clozapine-treated patients and it is estimated that clozapine treatment resistance reaches 10% of patients with schizophrenia [7]. Since clozapine began to be used to treat schizophrenia patients in the late 1980s, antipsychotic drugs that show better effect than clozapine have not been developed. This is due to the lack of understanding of the pathophysiology of mental disorders, the incomplete identification of molecular-pharmacological targets of existing drugs, the lack of preclinical models, and the factors that make it difficult to predict accurately the clinical response due to the diversity of disease courses [8-10]. To improve the symptoms of clozapine-resistant schizophrenia patients, additional pharmacological or biological treatments such as antidepressants, mood stabilizers, and electroconvulsive therapy are combined, but the effect is just temporary or insufficient to control the mental illness. Thus, a new treatment target and related drug discovery is needed to overcome schizophrenia.
Olanzapine and quetiapine are classified as thienobenzodiazepine and dibenzothiazepine, respectively. They have chemical structures similar to clozapine, which is classified as dibenzodiazepine (Figure 1) [11]. Quetiapine is used in the guideline as a first-line drug for schizophrenia and bipolar affective disorder. Olanzapine has stronger efficacy but also a strong weight-gain effect, so it is used as a second-line drug [12-14]. Like most small molecule drugs, clozapine, olanzapine, and quetiapine all have multiple targets. In particular, psychiatric drugs act as magic shotguns rather than magic bullets [9].
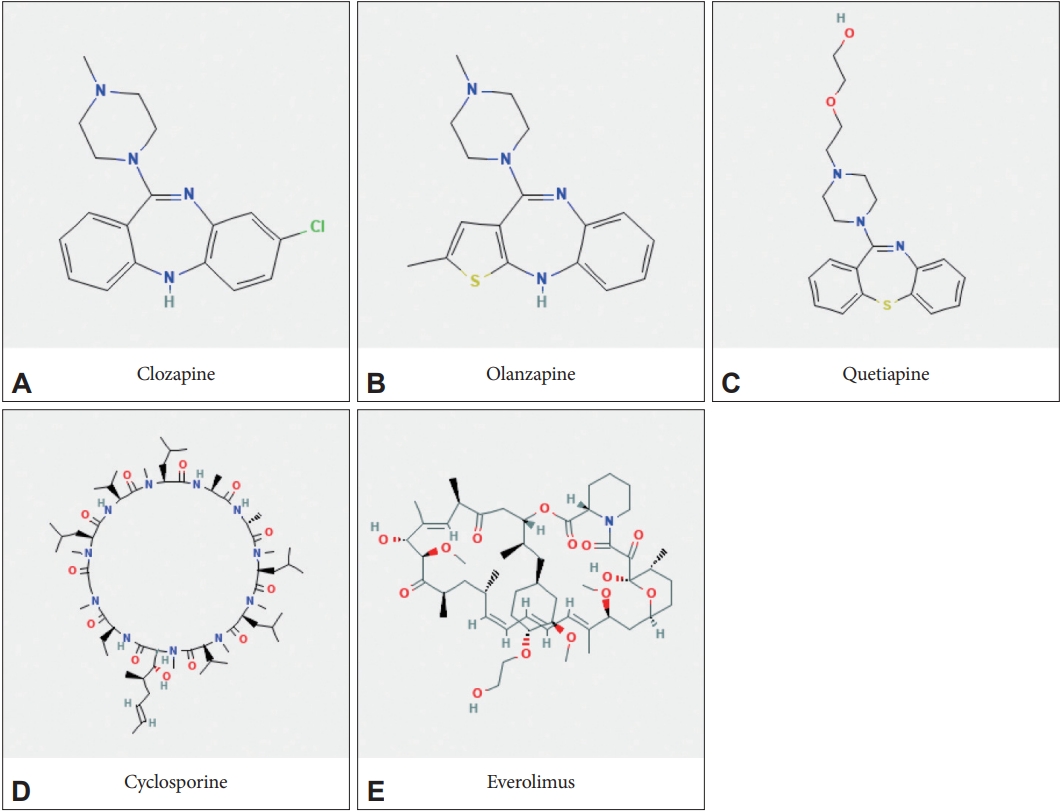
Chemical structures of antipsychotic and immunosuppressive drugs focused in deep learning-based molecular docking. A: Clozapine (the compound number in PubChem, 135398737). B: Olanzapine (135398745). C: Quetiapine (5002). D: Cyclosporin A (5284373). E: Everolimus (6442177).
Known targets of clozapine include 5-HT2A, 5-HT2C, D2, D4, alpha1, alpha2, H1, M1-5, N-methyl-D-aspartate (NMDA), and GABAB receptors [15,16]. Drugs such as Ulotaront, the TAAR1 agonistic and non-D2 antagonistic antipsychotic offers hope to treat schizophrenia [17,18]. However, overall, there are currently no antipsychotics for schizophrenia more effective than clozapine, and new antipsychotic drugs for clozapine-resistant patients are urgently needed.
In this study, GNINA, a pretrained convolutional neural network (CNN) scoring function, was used to calculate binding poses. It is based on learned representations of 3D proteinligand interactions. GNINA, a fork of Smina, outperforms Smina and AutoDock Vina in virtual screening methods [19,20]. Also in this study, the protein structure predicted by AlphaFold2 was used, especially the drug target receptors [21]. Through this study, receptors known to affect schizophrenia-related neurotransmitters, as well as immunological, metabolic, and neural stem cell growth were searched and found in the AlphaFold Protein Structure Database. Then they were analyzed with GNINA. The differences in receptor affinities according to drugs were measured, and receptors that could be used in drug discovery for schizophrenia are proposed for further study.
In detail, using the already developed GNINA and AlphaFold, the neuropharmacological, immune, and metabolic receptors that specifically act on each drug among clozapine, olanzapine, and quetiapine are explored. And based on these results, they discovered specific drugs that act on these receptors.
METHODS
Supplementary Table 1 (in the online-only Data Supplement) is a list of the receptors included in this study, which were classified as neuropharmacological (neuro), immunological (immune), or metabolic receptors [8,22-27]. In particular, it contains receptors involved in the metabolism of neural stem cells [27-30].
Ligands other than clozapine, olanzapine, and quetiapine in Supplementary Table 2 (in the online-only Data Supplement) were included in this study as the most commonly prescribed drugs [31-33] (including illegal drugs) and drugs used for targeted therapy [34]. Monoclonal antibodies were excluded because they could not be measured using GNINA.
The protein pdb file of the target receptor was downloaded from the AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk/). The downloading code was automated using the crawling library called selenium and request.
PDBFixer in OpenMM was used to perform the tasks that needed to be processed before performing docking simulation between protein and ligand [35]. In short, PDBFixer is an application that fixes problems in the Protein Data Bank files to prepare them for simulation.
DeepChem (https://deepchem.io/) makes the most suitable pose for protein and ligand docking. This prepares protein-ligand complexes for docking. Afterwards, using the processed pdb file and SMILES of the drug, we declared PoseGenerator in GNINA, and docked the protein and ligand with generate_poses in the default scoring setting. As a result, scores displayed the results obtained through a pretrained CNN, including binding affinity. The scores include binding affinities (kcal/mol), CNN pose scores (a probability that the pose has a low root mean square deviation to the binding pose), and CNN affinities predicted by GNINA19. The Pearson correlation efficient value between binding affinities and CNN affinities was -0.66, (p<0.0001). In this study, the results were compared using only CNN affinity values. We used the highest values of CNN affinities, and there was no difference in the order of receptors between the lowest and highest values.
The PyMOL Molecular Graphics System, Version 1.8 (Schrödinger LLC., New York, USA) was used to visualize the ligand-target interaction. No institutional review board approval is required as no humans or animals were involved in this study.
RESULTS
Top 20 CNN affinities by receptor
We obtained CNN binding affinities values for each of the 220 receptors that interacted with the three drugs via GNINA (Figure 2). The highest CNN affinities value was obtained from clozapine-bound cannabinoid receptor 2 (CB2, 7.522), olanzapine-bound serotonin 1B receptor (5-HT1BR, 7.004), olanzapine-bound muscarinic cholinergic receptor 5 receptor (M5, 6.713), and olanzapine-bound C-C chemokine receptor type 5 receptor (CCR5, 6.677) followed. Moreover, quetiapine showed the highest value with neuropeptide Y 4 receptor (NPYR4, 6.657). Overall, out of the top 20, 8 neuro receptors (red), 5 immune receptors (green), and 7 metabolic (blue) receptors were included in the list. Figure 2 shows the 3D structure of the receptor with the highest CNN affinities for each ligand.
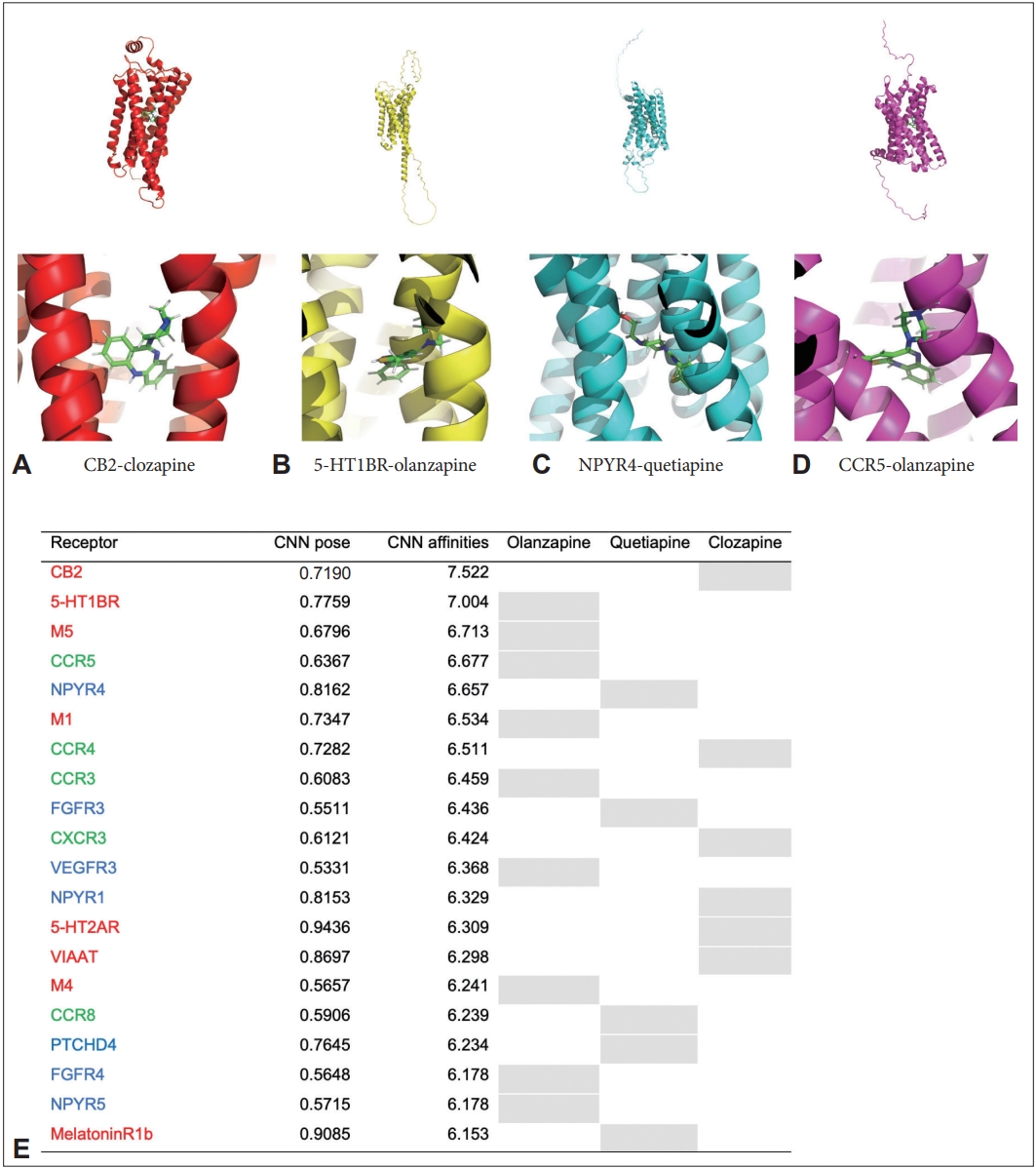
Top 20 convolutional neural network (CNN) affinities by receptor. A: A representative image of binding between cannabinoid receptor 2 (CB2) and clozapine. B: A representative image of binding between sertotonin receptor 1B (5-HT1BR) and olanzapine. C: A representative image of binding between neuropeptide Y receptor 4 (NPYR4) and quetiapine. D: A representative image of binding between C-C chemokine receptor 5 (CCR5) and olanzapine. E: Top 20 list of CNN affinities by receptor, neuropharmacological receptors are shown in red, immunological receptors in green, and metabolic receptors in blue. Shading in the table indicates the drug-receptor binding pair with the highest affinity. 5-HT, 5-hydroxytryptamine (serotonin) receptor; CB, cannabinoid receptor; CCR, C-C chemokine receptor; CXCR, C-X-C chemokine receptor; FGFR, fibroblast growth factor receptor; M, Muscarinic acetylcholine receptor; NPYR, neuropeptide Y receptor; PTCHD, patched domain-containing protein; VIAAT, vesicular inhibitory amino acid transporter.
CNN affinities among receptors for antipsychotics
Among the receptors bound to clozapine, CB2 had the highest value, followed by CCR4 (6.511), CCR5 (6.500), CCR3 (6.451), and C-X-C chemokine receptor 3 (CXCR3, 6.424). 4 neuro receptors, 10 immune receptors, and 6 metabolic receptors were included in the list for CNN affinities among receptors for clozapine, and immune receptors were the most common in the top 20. Keratin was included in the analysis as a control for the receptors.
Among the receptors bound to olanzapine, 5-HT1BR had the highest value, followed by M5 (6.713), CCR5 (6.677), M1 (6.534), and CCR3 (6.459). Neuro receptors were 5, immune receptors 4, metabolic receptors 11 in the list, and metabolic receptors were the most common.
Among the receptors bound to quetiapine, NPYR4 had the highest value, followed by fibroblast growth factor receptor 3 (FGFR3, 6.436), CCR5 (6.296), CCR8 (6.239), and patched domain containing 4 (PTCHD4, 6.234). Neuro receptors were 8, immune receptors 6, metabolic receptors 6 in the list, and metabolic receptors were the most common (Table 1).
CNN affinities among the receptors bound by the classified receptor system
In neuro receptors, the order of CNN affinities was CB2 (7.522), 5-HT1BR (7.004), M5 (6.713), M1 (6.534), and 5-HT2AR (6.309). More than two receptors related to 5-HT, muscarinic, CB, and melatonin were included, and TAAR, VIAAT, VGLUT1, and GRIK4 were also included.
Among immune receptors, CNN affinities of CCR5 (6.677), CCR4 (6.511), CCR3 (6.459), CXCR3 (6.424), and CCR8 (6.239) were shown. Two or more CCR, CXCR, and prostaglandin-related proteins were included, and the C5a receptor, toll-like receptor 2, and prostacyclin receptor were also included.
In the metabolic receptors, CNN affinities were shown in the order of NPYR4 (6.657), FGFR3 (6.436), vascular endothelial growth factor 3 (VEGFR3, 6.368), NPYR1 (6.329), and PTCHD4 (6.234). Two or more NPYR, orexin receptor, FGFR and melanocortin receptor-related proteins were included, and vasopressin receptor, oxytocin receptor, progesterone receptor, platelet-derived growth factor, Tropomyosin receptor kinase C receptor, and glial cell-derived neurotrophic factor were also included (Table 2).
Top 20 CNN affinities among widely used drugs and the highest binding receptors for antipsychotics
We found that CB2, 5-HT1BR, NPYR4, and CCR5 showed high affinities with the antipsychotics through this study, so we performed additional analysis to search for affinities with a wider range of ligands for these receptors.
Surprisingly, cyclosporine showed the highest affinities for all three receptors except for CCR5 (CB2 7.996, 5-HT1BR 7.423, NPYR4 7,785), everolimus had the highest affinities for CCR5 (7.584), and the other three receptors had the second highest affinities with it (CB2 7.388, 5-HT1BR 7.422, NPYR4 7.420).
In addition, for CB2: octreotide (7.120), trametinib (7.036), and maraviroc (6.979) showed high affinities (in this order), and for 5-HT1BR: salinomycin (7.105), tacrolimus (7.050), nilotinib (6.913), and for NPYR4: crizotinib (7.100), nilotinib (6.976), lapatinib (6.953), and for CCR5: nilotinib (7.420), crizotinib (7.417), maraviroc (7.410), and cyclosporine (7.373) were found in that order (Figure 3).
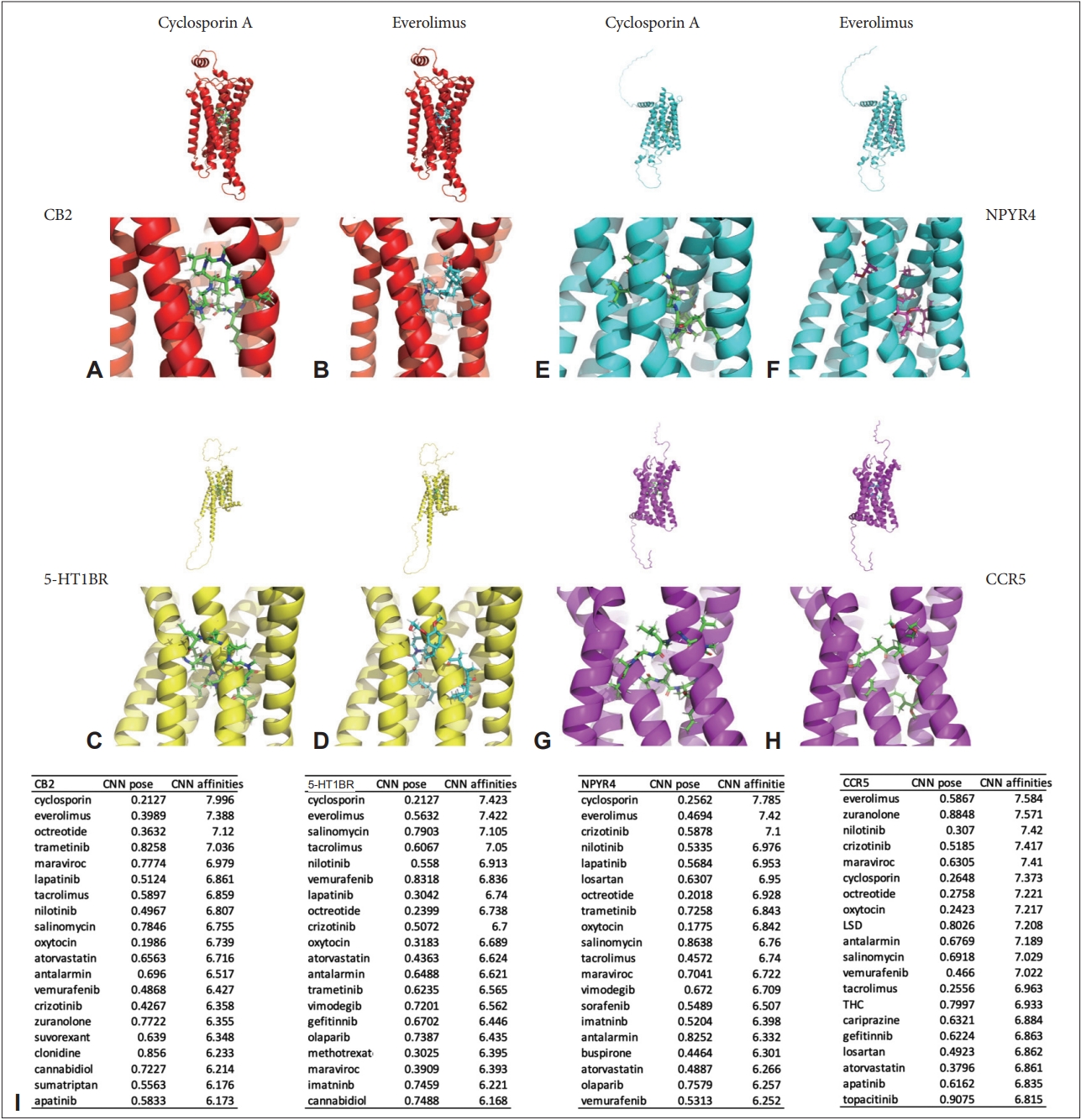
Top 20 convolutional neural network (CNN) affinities among widely used drugs and the highest binding receptors for antipsychotics. A: A representative image of binding between cannabinoid receptor 2 (CB2) and cyclosporin A. B: A representative image of binding between CB2 and everolimus. C: A representative image of binding between sertotonin receptor 1B (5-HT1BR) and cyclosporin A. D: A representative image of binding between 5-HT1BR and everolimus. E: A representative image of binding between neuropeptide Y receptor 4 (NPYR4) and cyclosporin A. F: A representative image of binding between NPYR4 and everolimus. G: A representative image of binding between C-C chemokine receptor 5 (CCR5) and cyclosporin A. H: A representative image of binding between CCR5 and everolimus. I: Top 20 list of CNN affinities by receptors found in this study including CB2, 5-HT1BR, NPYR4, and CCR5, neuropharmacological receptors are shown in red, immunological receptors in green, and metabolic receptors in blue. LSD, lysergic acid diethylamide; THC, tetrahydrocannabinol.
DISCUSSION
Throughout this study, we used CNN-based GNINA to estimate the affinities of clozapine, olanzapine, and quetiapine with neuropharmacological, immunological, and metabolic receptors predicted from AlphaFold. Through this in silico study, we confirmed that antipsychotic drugs act not only on neuropharmacological receptors but also on immune and metabolic receptors. In particular, CB2 for clozapine, 5-HT1BR for olanzapine, and NPYR4 for quetiapine showed the highest affinities.
Neurons and glia in the brain are expected to account for about 100 billion cells, respectively [36]. In addition to the rapid electrophysiological properties of neurons, which are excitable cells, the modulatory effect of G protein-coupled receptor is also important for cellular functions, including signal transduction [37,38]. The function of neurons cannot be maintained without the role of the glia cells that make up the brain. Therefore, it is essential to understand the action of antipsychotics on immunological and metabolic receptors as well as neuropharmacological receptors. In this study, we performed an exploratory study involving a variety of receptors to understand the interactions of neurons and glia and the functioning of the brain within the body’s immune and metabolic environment.
In the field of psychiatry, drug discovery including virtual screening is actively progressing, but there are still unmet needs and it is one of the most difficult areas to develop new drugs [39,40]. However, looking at some studies using deep learning and virtual screening, there is a study that predict drug repositioning using various deep learning techniques in psychiatric disorder including schizophrenia [41]. In addition, Another study utilized deep learning techniques to structurally predict the GABA receptor action of herbal compounds [42]. Finally, one study conducted to discover optimized NMDA receptor antagonists for Alzheimer’s disease using Smina and idock for virtual screening [43].
In a previous study of CB2, nabilone-induced exacerbation of psychotic symptoms was reported in patients with Parkinson’s disease [44]. Nabilone is a synthetic form of tetrahydrocannabinol for nausea and vomiting in cancer patients and HIV patients; it acts as a partial agonist for both CB1 and CB2. The most worrisome side effect is psychotic symptoms, but only two cases have been reported in studies targeting post-traumatic stress disorder patients [45]. Unlike CB1, which is distributed in the central nervous system, CB2 is distributed in the peripheral nervous system and also in immune cells such as lymphoid tissue and microglia [46]. The antibiotic minocycline acts on CB2 and is expected to serve as an adjuvant treatment to prevent the onset or exacerbation of schizophrenia [47-49].
Previous studies on 5-HT1BR have shown that triptan drugs, such as sumatriptan for migraine treatment, act on receptors such as 5-HT1AR, 5-HT1DR, 5-HT1FR, and 5-HT1BR [50]. The action of 5-HT1BR as a selective serotonin reuptake inhibitor is also reported [51]. Increased 5-HT1BR messenger ribonucleic acid in the dorsolateral prefronal cortex tissue [52], and an increase in 5-HT1BR mRNA in the hippocampus were also reported in a schizophrenia biomarker study [53].
A previous study on NPYR4 showed that obinepitide was an investigational drug acting on NPYR4, and it showed weight loss in experimental animals by acting on NPYR2 and NPYR4 [54]. Because of the high comorbidity of weight gain and metabolic syndrome due to schizophrenia itself and antipsychotics, it seems to have both the implications of a therapeutic target and a receptor associated with drug side effects [55]. NPYR4 has not been studied for schizophrenia, thus this study suggests a new receptor target.
CCR5 is a chemokine receptor for regulated on activation, normal T cell expressed and secreted as its ligand and is known to be expressed in glia and neural stem cells [56]. Because CCR5 is known as the infection route of HIV 1, maraviroc (which inhibits CCR5) has been developed and used [57]. Moreover, there is no study about treating schizophrenia using maraviroc. In a CCR5 gene deletion animal study, CCR5-deleted mice showed a decrease in social recognition [58]. In a schizophrenia genome study, higher expression of CCR5 was reported compared to the control [59], and the frequencies of CCR5-A55029GA genotypes and CCR5-A55029GAG genotypes were found to be higher in patients. In a genomic study of 268 schizophrenia and 323 controls, association with the CCR5 32 deletion allele was reported in the late onset schizophrenia group [60]. After combining previous studies, it is now expected that the regulation of CCR5 may affect the treatment of patients with schizophrenia.
Cyclosporine is a calcineurin inhibitor mainly used as an immunosuppressant. Cyclosporine-induced neurotoxicity is a well-known side effect and, in particular, can induce psychosis [61-65]. Everolimus, also an immunosuppressant, is a drug that acts on the mechanistic target of rapamycin and that affects the growth of neural stem cells [66]. A case study has been reported in which everolimus also induced psychosis [67], but in a previous study, the use of everolimus in children and adolescents with tuberous sclerosis resulted in a decrease in prepulse inhibition, an indicator of psychosis and sensorimotor gating [68-70]. In this study, high binding affinities between cyclosporine and everolimus, and the receptors found in this study, were discovered. Now, a study to develop drugs that bind to the corresponding receptors, but have the opposite action through pharmacodynamics, can be considered.
In this study, we evaluated only CNN affinity by ligand-receptor binding in silico, and evaluation of the pharmacodynamic effect for function is needed in the future. Moreover, physical measurements of ligand-receptor binding, such as provided by x-ray crystallography, are also required for a new antipsychotics discovery. Using CNN-based GNINA, we could perform a more accurate deep learning-based analysis than with the existing AutoDock Vina. In addition, a wide range of receptors were searched, and in particular, the limitations of existing protein structure studies were supplemented using AlphaFold. By discovering a target for new antipsychotics that had not been studied before (such as NPYR4), we got an opportunity to provide new insight into the field of drug discovery for schizophrenia.
This study investigated olanzapine and quetiapine, which have chemical structures similar to those of clozapine, and which are the most effective drugs for controlling psychotic symptoms of schizophrenia. Furthermore, receptors upon which each drug acts were searched using GNINA for structure-based drug discovery (SBDD). It was confirmed that antipsychotics have high affinities on receptors (such as CB2 and CCR5) that are present in immune cells such as microglia, which exist in the immune part of the central nervous system. The receptors described in this study are suggested to play important roles in the improvement of psychiatric symptoms and the prevention of recurrence, and furthermore, in the pathophysiology of schizophrenia. Cyclosporin A and everolimus, which bind strongly to these receptors, have potential applications in the development of novel antipsychotic drugs.
Clozapine, olanzapine, and quetiapine have been studied mainly on serotonin and dopamine receptors in previous studies because of limitations in biological resources available in the past (structure of receptor proteins, accessibility, measurable technology, etc.) [71]. And there is a high need to explore the mechanism of drugs on a wider range of targets including receptor proteins. In addition, the relationship between immunological and metabolic abnormalities and psychosis was recently revealed [72,73]. Therefore, the effect on immunological and metabolic receptors could show therapeutic effects, and finding new receptors with new mechanisms and drugs specific to those receptors is expected to lead to the discovery of treatments for antipsychotic-resistant schizophrenia patients.
Although the mechanism of mental illness is still unknown, the therapeutic effect of antipsychotic drugs is obvious. Therefore, it is considered as a useful approach to explore the receptor binding of antipsychotic drugs and use them to search for new targets. We hope that the method used in this study for SBDD and drug repositioning for the development of new antipsychotics will be of great help to schizophrenia patients.
Supplementary Materials
The online-only Data Supplement is available with this article at https://doi.org/10.30773/pi.2022.0343.
List of receptors classified by system
List of ligands: the most commonly prescribed drugs and drugs used for targeted therapy
Notes
Availability of Data and Material
All data generated or analyzed during the study are included in this published article (and its supplementary information files).
Conflicts of Interest
The authors have no potential conflicts of interest to disclose.
Author Contributions
Conceptualization: Yangsik Kim. Formal analysis: Yangsik Kim. Funding acquisition: Yangsik Kim. Methodology: Seyong Kim. Resources: Seyong Kim. Software: Seyong Kim. Supervision: Yangsik Kim. Visualization: Yangsik Kim. Writing—original draft: Yangsik Kim. Writing—review & editing: Yangsik Kim.
Funding Statement
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (NRF-2021R1C1C1003266 to Y.K.), and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI22C0492 to Y.K.).


