Genetic Association Study of the Alpha 7 Nicotinic Receptor (CHRNA7) with the Development of Schizophrenia and Bipolar Disorder in Korean Population
Article information
Abstract
Objective
CHRNA7 has been shown to be a strong candidate gene for schizophrenia and bipolar disorder. It is located on chromosome 15q13-q14, which is one of the replicated linkage spots for schizophrenia and bipolar disorder.
Methods
We conducted an association study to determine whether previous positive association is replicable in the Korean population. We included 254 patients with schizophrenia, 193 patients with bipolar disorder type I, 38 patients with bipolar disorder type II, 64 schizoaffective disorder patients, and 349 controls. All subjects were ethnically Korean. A total of 898 subjects were included, and genotyping was done for three single nucleotide polymorphisms (SNPs) of CHRNA7. These three intronic SNPs were rs2337506 (A/G), rs6494223 (C/T), and rs12916879 (A/G).
Results
There was only one marginally significant association; this association was between rs12916879 and bipolar disorder type I in the male subgroup. In both the allele and genotype distributions, we found a weak signal (Chi-squared=3.57, df=1, p=0.06 for allele, Chi-squared=7.50, df=2, p=0.02 for genotype) only. Unphased haplotype analysis could not provide additional support for this finding. No SNP was associated with schizophrenia or any other affected groups in this Korean sample. The associative finding is marginal and inconclusive.
Conclusion
We could not replicate positive association in other ethnic groups previously studied. This suggests possible heterogeneity in the genes associated with schizophrenia and bipolar disorders. Because of structural complexity of the CHRNA7 gene and the limited statistical power of this study, further genetic studies with more SNPs and larger samples covering various populations, along with more fine molecular exploration of the CHRNA7 gene structure, are required.
INTRODUCTION
It has been observed that patients with schizophrenia are easily distracted by trivial sensory stimuli from the surroundings. This may represent disturbances in the filtering mechanism for sensory stimuli in the central nervous system.5,6 One auditory-evoked potential, P50, can reflect this kind of physiological abnormality in sensory gaiting. P50 measures the electrical brain wave with a 50-millisecond latency following paired auditory stimuli delivered 0.5 seconds apart.4 If normal sensory gating has occurred, the amplitude of the second response decreases compared with the response to the first stimulus. Patients with schizophrenia showed no decrease for the second stimulus and a greater response than that expected in the normal population.2,16 Similarly, bipolar patients with psychotic features also showed diminished suppression of the P50 auditory-evoked potential.34 Abnormal sensory gating has been found to be inherited. Reduced attenuation of the P50 response was reported for the first-degree of relatives of individuals with schizophrenia, and for the relatives of individuals with bipolar disorder with psychotic features.17,38 An abnormality in P50 was observed in mice with a dissected fimbria-fornix, and the abnormality was normalized with nicotine. In patients with schizophrenia, abnormal P50 was transiently normalized with nicotine use.1,25 These findings are consistent with the fact that schizophrenics are more likely to be smokers than normal individuals. The intake of nicotine associated with smoking could be a self-compensation for neuro-physiological deficits.20
CHRNA7 has been considered a strong candidate gene for schizophrenia because of its function and its chromosomal location. Abnormal activity of the CHRNA7 receptor is responsible for P50 deficits. In animal models, pharmacological antagonists of the CHRNA7 receptor induce the inhibitory deficit of P50.30 Several studies have found decreased expression of the CHRNA7 gene in the postmortem human hippocampus, reticular thalamic nucleus and frontal cortex of schizophrenic subjects.8,18,21 CHRNA7 gene encodes a single peptide consisting of an N-terminal extracellular domain, three membrane-spanning regions, an intracellular domain, a fourth transmembrane region, and an extracellular C-terminal tail. The CHRNA7 gene contains 10 exons and spans more than 75 kb.19
The CHRNA7 gene is located on chromosome 15q13-q14. Freedman et al.17 reported a lod score of 5.3 at a marker (D15S 1360) on intron 2 of the CHRNA7 gene in a linkage study for P50. Since then, there have been many linkage studies focusing on this region. The chromosome 15q13-q14 region itself is a strong linkage spot for both schizophrenia and bipolar disorder. Many positive linkage findings, as well as several negative studies, exist for schizophrenia in this chromosomal region.26 One meta-analysis for schizophrenia supported positive linkage association,3 but another meta-analysis did not.28 For bipolar disorder, linkage disequilibrium at D15S1360 and linkage at 15q13-q14 have been reported.11,32 Age at onset and a good response to lithium also have been mapped to this region.13,39 Within the CHRNA7 gene, many single nucleotide polymorphisms (SNPs) in exons, introns and promoter sites have been tested for association with schizophrenia and bipolar disorder, and these experiments have produced conflicting results.22,26,27
There has been no genetic study on the CHRNA7 gene and schizophrenia or bipolar disorder for the Korean population. We conducted this study in order to further explore the role of the CHRNA7 gene by attempting to replicate previous positive association between the CHRNA7 gene and schizophrenia and bipolar disorder in a Korean sample.
METHODS
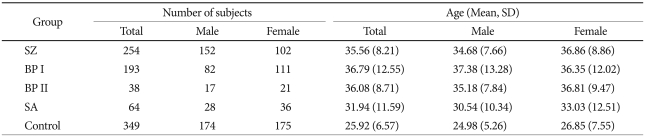
Patients were recruited from the Seoul National University Hospital and several psychiatric clinics in Korea. All patients satisfied the diagnostic criteria of DSM-IV for schizophrenia and bipolar disorder. They were individually interviewed by trained nurses using the Korean version of Diagnostic Interview for Genetic Studies.24,33 Consensus diagnostic meetings of more than three psychiatrists were held regularly to make the participants' final diagnoses. Subjects with a history of any kind of organic abnormality of the brain, substance dependence, drug abuse, or other physical conditions possibly manifesting psychiatric symptoms were excluded from this study. The final analyses were performed with 549 patients and 349 controls. The patient group consisted of 254 patients with schizophrenia, 193 patients with bipolar disorder type I, 38 patients with bipolar disorder type II, and 64 patients with schizoaffective disorder. Healthy controls were free from present, past and family history (first-degree relatives) of psychiatric illness or substance abuse diagnoses. The total number of control subjects was 349 (174 males and 175 females). The mean ages of the patient and control groups were 35.09±10.27 and 25.92±6.57 years, respectively. More detailed information regarding subject number and mean age for each diagnostic group is given in Table 1. All subjects participating in this study signed a written informed consent form, and the study protocol was approved by the ethics committee of Seoul National University Hospital.
Deoxyribonucleic acid (DNA) was extracted from blood samples using a DNA isolation kit (Roche, Mannheim, Germany). We selected three intronic SNPs for the CHRNA7 gene: rs2337506, rs6494223, and rs12916879. These SNPs were selected based on chromosomal location, minor allele frequencies in Asians, and linkage disequilibrium (LD), according to public information such as Entrez SNP of National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/snp). Even though the number of SNPs was so limited to cover the whole CHRNA7 gene, we tried to include representative SNPs of the whole gene and choose the closer SNP to the markers previously studied for schizophrenia and bipolar disorder. Rs2337506 (chr15:32,345,817-32,346,317bp) is located inside intron 2, rs6494223 (chr15:32,396,207-32,396,707bp) is located in intron 3, and rs12916879 is located in intron 4 (chr15:32,417,083-32,417,583bp). According to the haplotype information in Caucasians, these three SNPs are in separate haplotype blocks.7 The D15S1360 microsatellite marker, which is the most frequently studied and reported to be associated with schizophrenia and bipolar disorder, is located in intron 2. Therefore SNP rs2337506 is close to D15 S1360 (chr15:30,143,950-30,144,057) and is likely to be in LD. Genotyping was performed using the TaqMan™ method (Applied Biosystems, Foster City, CA, USA).31 The chromosomal location of SNPs were adapted from UCSC Genome Browser on Human Feb. 2009 (GRCh37/hg19) Assembly (http://genome.ucsc.edu/).
Hardy-Weinberg equilibrium was tested by goodness-of-fit chi-squared test. Contingency chi-squared tests were performed for the comparison of allele and genotype frequencies between the patients and controls using Statistical Package for the Social Sciences (SPSS)(version 13.0; SPSS Inc., Chicago, IL, USA). The haplotype analyses were done using UNPHASED 3.0.13 (http://www.mrc-bsu.cam.ac.uk/personal/frank/software/unphased/).11 The haplotype association was tested for two to three SNPs with the sliding window method. In this study, the program calculated chi-squared values and p-values for a specific haplotype association according to the main haplotype effect model. The level of statistical significance was set at p=0.05. Genetic Power Calculator (http://pngu.mgh.harvard.edu/~purcell/gpc)35 was used to compute the statistical power of our sample.
RESULTS
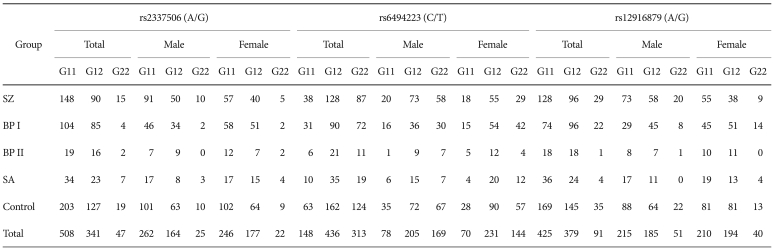
The allele and genotype distributions in both the whole patient and control groups were in Hardy-Weinberg equilibrium for the three selected SNPs (data not shown). The whole patient group included the patients with schizophrenia, bipolar disorder type I and type II, and schizoaffective disorder. For the analysis, all patients were regrouped into several affected groups: a group of patients with schizophrenia, a group of patients with bipolar disorder type I only, a group of patients with bipolar disorder type I and type II, a group of patients with bipolar disorder type I, II and schizoaffective disorder, and a group of all the patients. However, in the tables, the raw data are shown according to each diagnostic group and not according to the testing group in order to provide the exact count of alleles and genotypes for each diagnostic group (Table 2 and 3).
There was no allelic association found for either schizophrenia or bipolar disorder. Additional analysis according to sex did not find any association for any of the three SNPs. Only one marginally significant association was found; this association was between rs12916879 and bipolar disorder type I in the male subgroup (Chi-squared=3.57, df=1, p=0.06). Table 2 shows the allele distribution for the three SNPs of the CHRNA7 gene for each diagnostic group. The genotype distribution for each diagnostic group is presented in Table 3. In the total sample (male and female), no significant association with genotype distribution was observed for any of the SNPs. When we divided the groups by sex, rs12916879 was significantly associated with bipolar disorder type I (p=0.02) only in males. The degree of significance was reduced if the analysis was extended to include bipolar disorder type II and schizoaffective disorder (BPD type I & II, p=0.05, BPD type I, II & SA, p=0.05). There was no genotypic association for any of the three SNPs in females for any diagnostic group. No SNP was associated with schizophrenia or any other affected groups in this sample.
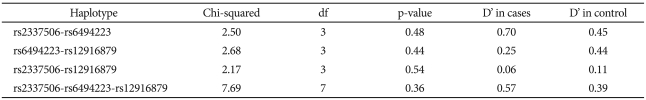
Since a positive association with a single SNP was found only in males, further haplotype analysis was performed only for males. Table 4 summarizes the global D' and association statistics between SNPs. No haplotype was significantly associated with bipolar disorder. In the effort to reduce the possible influence of multiple testing, a permutation test was performed using the UNPHASED genetic analysis software. One thousand permutations were conducted for the male bipolar disorder type I subgroup and for controls. The adjusted p-value from the permutation test was 0.28 (standard error=0.01).
DISCUSSION
We found a positive genotypic association between bipolar disorder type I and SNP rs12916879 in the male subgroup. However, the association was not supported by allele or haplotype, and the significance could not survive correction for multiple testing. For schizophrenia, we could not find any association with CHRNA7 in this Korean sample. As a whole, we could not replicate the positive association of CHRNA7 with either schizophrenia or bipolar disorder in this Korean sample. This association study does not support a strong genetic role of CHRNA7 in schizophrenia and bipolar disorder.
This result indirectly supports the well-known genetic heterogeneity among various populations for both bipolar disorder and schizophrenia. There are many positive linkage and association findings for this chromosomal region in various ethnic populations.26 However, these associations have not been supported in Asian populations. There has been a report of a positive linkage for Han Chinese patients between schizophrenia and the chromosome 15q14 region.29 However, two negative association studies in the Han Chinese population have been published so far.12,23 To the best of our knowledge, there has been no published result on Japanese patients with schizophrenia.
It is well known that there is a difference in allele frequencies between various ethnic populations. In this Korean sample, the allele frequencies of three SNPs of the CHRNA7 gene were similar to the allele frequencies of Han Chinese and Japanese populations reported in Entrez SNP: A in SNP rs2337506 (0.76) is 0.66 for Japanese and 0.77 for Han Chinese, T in SNP rs6494223 (0.59) is 0.76 for Japanese and 0.67 for Han Chinese, and A in SNP rs12916879 (0.69) is 0.64 for Japanese and 0.71 for Han Chinese.
Chromosome 15q13-14, as well as the CHRNA7 gene, has a complex structure. Exons 5 to 10 of CHRNA7 are known to be duplicated and located in the opposite direction. This duplication and inversion creates CHRFAM7A, an alpha 7-like nicotinic receptor subunit gene. This gene is a hybrid of CHRNA7 and several copies of an unrelated genetic sequence FAM7A.15 The duplication, a type of copy number variant, is not present in all subjects. About 30% of individuals have only one copy, and more than 5% have no copies.19,37 There is 2-bp deletion polymorphism in exon 6 of CHRFAM7A, which is tightly linked to the duplication-inversion polymorphism.15 Previous studies using this 2-bp deletion polymorphism as a surrogate marker for the copy number variant of CHRNA7 have been conducted. In French Caucasians, this 2-bp deletion was a risk factor for P50 sensory gating deficits.36 Flomen et al.14 could not find an association between this 2-bp deletion polymorphism and schizophrenia or bipolar disorder in the Scottish population. The presence of the 2-bp deletion was associated with poor performance in episodic memory tests in the family members of individuals with schizophrenia.9 This kind of complex genetic structure probably contributes to the discrepancies in the current linkage and/or association results for psychiatric disorders. The three SNPs of this study are located in intron 2, 3 and 4, not in duplicated region of the gene. Unfortunately, we could not examine this 2-bp deletion polymorphism. Since the 2-bp deletion strongly represents the complicated structure of CHRNA7, further studies that include this polymorphism will be crucial to understand the role of CHRNA7 as well as the whole chromosomal region of 15q13-14.
This study has considerable limitations. First, the sample size was not large enough to detect small gene effects. Considering the minor allele as a risk allele and given the significance level of α=0.05, our analysis had a power of 47-61% for schizophrenia, and 41-55% for bipolar disorder type I under a dominant model (genotypic relative risk of AA=1.5; prevalence=0.01 for both schizophrenia and bipolar disorder type I). Therefore, we admit that our study sample was limited in statistical power, even though this sample size was not relatively small. Second, we included only three SNPs of CHRNA7, which do not represent the whole CHRNA7 gene. Third, this study did not include SNPs in the promoter site or the 2-bp deletion in exon 6, both of which have been reported to have meaningful associations in other population samples. Fourth, our control group was significantly younger than the patient group, and we could not exclude the possibility of future development of schizophrenia and/or bipolar disorder among this control group. Finally, the population stratification should be kept in mind for every association study, even though the Korean population is considered relatively less heterogeneous.
CHRNA7, which is located on chromosome 15q13-14, has a complex genetic structure and has been frequently implicated in many psychiatric illnesses. Because there are remarkable inconsistencies in the results from previous studies, and because many of them had insufficient statistical power, it is necessary to conduct further research on the CHRNA7 gene with respect to schizophrenia and bipolar disorder with sufficient statistical power in many other populations. We hope the data of this study will contribute to the future meta-analysis, although the power of this study alone is not enough to detect any genetic association.
Acknowledgments
This work was supported by National Research Foundation of Korea Grant funded by the Korean Government (2010-0015782 & KRF-2008-531-E00045).