Polymorphism of Estrogen Receptor Genes and Its Interactions With Neurodevelopmental Genes in Attention Deficit Hyperactivity Disorder Among Chinese Han Descent
Article information
Abstract
Objective
Attention deficit hyperactivity disorder (ADHD) is a polygenic neurodevelopmental disorder with significant gender differences. The sexual dimorphism of ADHD may be associated with estrogen acting through estrogen receptors (ESR). This study investigates the impact of ESR gene polymorphism and its interactions with neurodevelopmental genes on ADHD susceptibility.
Methods
The study compared genotyping data of single nucleotide polymorphisms in ESR1 and ESR2 in 1,035 ADHD cases and 962 controls. The gene-gene interactions between ESR genes and three neurodevelopmental genes (brain-derived neurotrophic factor [BDNF], synaptosomal-associated protein of 25 kDa gene [SNAP25], and cadherin-13 [CDH13]) in ADHD were investigated using generalized multifactor dimensionality reduction and verified by logistic regression analysis.
Results
The G allele of rs960070/ESR2 (empirical p=0.0076) and the A allele of rs8017441/ESR2 (empirical p=0.0426) were found significantly higher in ADHD cases than in the controls but not in male or female subgroups. Though no difference was found in all subjects or females, the A allele of rs9340817/ESR1 (empirical p=0.0344) was found significantly higher in ADHD cases than controls in males. We also found genetic interaction models between ESR2 gene, neurodevelopmental genes and ADHD susceptibility in males (ESR2 rs960070/BDNF rs6265/BDNF rs2049046/SNAP25 rs362987/CDH13 rs6565113) and females (ESR2 rs960070/BDNF rs6265/BDNF rs2049046) separately, though it was negative in overall subjects.
Conclusion
The ESR gene polymorphism associates with ADHD among Chinese Han children, with interactions between ESR genes and neurodevelopmental genes potentially influencing the susceptibility of ADHD.
INTRODUCTION
Attention deficit hyperactivity disorder (ADHD) is a common childhood neurodevelopmental disorder, affecting 5%– 11% of children [1,2]. Heritability estimates for the disorder are Print ISSN 1738-3684 / On-line ISSN 1976-3026 OPEN ACCESS approximately 74% [3], indicating a strong genetic component. Previous studies have further demonstrated that ADHD is polygenic, influenced by the combined effects of multiple genes, each with a minor influence [4]. Simultaneously, ADHD significantly varies across sex. Epidemiological studies have observed an obvious gender dimorphism in ADHD, with a male to female ratio of approximately 3-4:1 [5]. Significant sex differences in clinical symptoms, brain structure and function have also been reported in numerous studies of ADHD patients [6]. Researchers have previously explained the gender differences in ADHD from three aspects: biological factors, sociocultural factors, and gender bias in diagnostic. Research on the genetic mechanism of sex dimorphism in ADHD has mainly focused on X or Y chromosome genes, with comparatively less attention given to genes related to sex hormones.
Estrogen, which plays an important role in establishing neural and behavioral sex differences, has been suggested to be related to the sexual dimorphism of ADHD by some scholars [7,8]. It has been reported that estrogen can stimulate the growth of spinous cells and synapse, while also increase the expression of brain-derived neurotrophic factor (BDNF) [9,10]. The biological effects of estrogen are primarily mediated by the estrogen receptors (ESRs), including ESR1 and ESR2. ESRs possess neuroprotective and anti-inflammatory properties which may underlie their observed psychotropic effects [11]. Previous studies have indicated that ESRs can activate the PI3K/Akt neuroprotective signaling pathway in the brain and reduce neuronal apoptosis [12,13]. Polymorphism of ESR genes has been reported to be associated with a variety of psychiatric disorders. The single nucleotide polymorphisms (SNPs) rs2234693 and rs9340799 in ESR1 were found to be associated with the pathogenesis of schizophrenia [14,15]. ESR1 rs22346939 and rs9340799 polymorphisms were also found to be related to the risk of major depressive disorder [16]. In autism, rs1155819 in ESR1 and rs1152582 in ESR2 were reported to be associated with severities in the impairment of social interaction and emotional regulation [17]. Since ADHD shares similar phenotypes and underlying genetic basis with these disorders, polymorphisms of ESR genes may also play a role in the pathology of ADHD. However, few studies have explored the impact of ESR gene polymorphisms on ADHD.
Neurodevelopmental genes have been widely investigated in ADHD [18,19]. Among these, BDNF, synaptosomal-associated protein of 25 kDa gene (SNAP25), and cadherin-13 (CDH13) show sexual dimorphism and are closely related to ADHD [20,21]. BDNF promotes neuronal growth and differentiation, participating in neuronal plasticity [22]. It contains an estrogen-like response element that can be modulated by estrogen through activating the ESR, regulating its expression and thereby regulating neural activity [23]. Our previous study in 325 Han Chinese samples revealed a sex-specific association between BDNF Val66Met (rs6265) and ADHD [24]. In mice experiments, an ESR2 agonist significantly increased BDNF-TrkB signaling pathways and downstream cascades related to synaptic plasticity [25]. SNAP25, implicated in neurotransmission and neuroplasticity, was also found as an ADHD susceptibility gene. Feng et al. [26] reported that SNAP25 rs362987 polymorphisms significantly increase ADHD risk. Sex differences in altered stress responses were reported in SNAP25 mutant mice [27]. Similarly, some CDH13 SNPs, which negative regulator of axon growth during neural differentiation, were revealed to contribute ADHD pathophysiology, particularly rs6565113 [28]. CDH13 mutant mice exposed to early-life stress showed sex differences, with increased locomotor activity in females but unchanged impulsivity and activity in males [29]. It remains unclear whether the observed sexual dimorphism in the above analyses also involves the ESR genes.
The current study aims to explore whether ESR gene polymorphisms are associated with ADHD and investigate whether genetic interactions among ESR genes and the neurodevelopmental genes (BDNF, SNAP25, and CDH13) contribute to ADHD susceptibility.
METHODS
Subjects
A total of 1,035 children with ADHD were recruited from the child psychiatry clinic at Peking University Sixth Hospital, including 162 females and 873 males. ADHD diagnosis was made according to Diagnostic and Statistical Manual of Mental Disorders, 4th edition (DSM-IV) criteria using the Clinical Diagnostic Interview Scale based on a semi-structured interview conducted by experienced child psychiatrists. The inclusion criteria were: 1) diagnosed as ADHD according to DSMIV, 2) age between 6 and 16 years, 3) full-scale intelligence quotient ≥70, 4) both biological parents were of Chinese Han descent, and 5) never used any psychiatric medication. Children with previous or present childhood schizophrenia, bipolar disorder, autism, intellectual disability, epilepsy, or other mental illness were excluded. Patients with obvious somatic or neurologic abnormalities, consciousness disorders, and current dependence or abuse of substance were also excluded. A total of 962 control samples of Chinese Han descent were recruited, including 355 females and 607 males. Controls included both children and adults and were recruited from three sources: local elementary school students, healthy blood donors attending the Blood Center of Peking University First Hospital, and healthy volunteers from our institute. The parents or adults completed the ADHD Rating Scale IV to exclude ADHD. Major psychiatric disorders, family history of psychosis, severe physical diseases, and substance abuse were also excluded [30].
This study adhered to the guidelines of the Declaration of Helsinki and was approved by the Ethics Review Committee of Peking University Sixth Hospital. All subjects or parents/guardians of children signed an informed consent form to participate. The Institutional Review Board (IRB) number is PKUH6 (2007)-(39).
Genotyping and SNPs selection
Genomic DNA was extracted from peripheral blood samples of cases and controls using standard protocols. SNP genotyping was performed using the Affymetrix6.0 array at CapitalBio Ltd. (Beijing, China) and following the Affymetrix standard protocol. A total of 131 SNPs in ESR1 and 26 SNPs in ESR2 were extracted. Haploview software version 4.2 (http://www.broad.mit.edu/mpg/haploview/) was used to select SNPs with minor allele frequency (MAF) >5% and meeting the Hardy–Weinberg equilibrium (HWE), yielding 97 ESR1 SNPs and 21 ESR2 SNPs. Linkage disequilibrium was identified using the confidence interval (CI) method in Haploview, and tag SNPs with a threshold of r2>0.80 were selected for subsequent analysis. Finally, 20 SNPs (ESR1: 15 SNPs; ESR2: 5 SNPs) were chosen as tag SNPs (Table 1).
SNPs for neurodevelopmental genes were selected based primarily on the previous studies. The BDNF SNPs, rs6265 [24,31], rs2030324 [32], and rs7934165 [33] have been studied in ADHD. The SNAP25 SNP rs362987 [26] and CDH13 SNP rs6565113 [34] were also associated with ADHD. Table 1 demonstrated the HWE test results and MAF information for the SNPs of ESR genes, BDNF gene, SNAP25 gene, and CDH13 gene that included in the final analysis. All the SNPs were with a HWE p-value and MAF over 0.05.
Statistical analysis
Statistical analysis of clinical data
Descriptive statistics were performed on demographic and clinical variables. Continuous variables with a normal distribution were expressed as mean±standard deviation, skewed continuous variables as median (interquartile range). Student’s t-test was used for two-group comparison, and the Mann-Whitney U test was used when the data did not conform to normal distribution. Categorical variables were expressed as numbers (percentages) and compared with the chi-square test. A p<0.05 was considered statistically significant. This part of the analysis was done using IBM SPSS 21 (IBM Corp., Armonk, NY, USA).
Case/control association test for SNPs
We used Haploview version 4.2 to perform HWE test and MAF of SNPs to ensure data quality. HWE was calculated using the exact test developed by Wigginton et al. [35] that implemented in Haploview. Haploview was also used to identify linkage disequilibrium. The linkage disequilibrium (LD) patterns of ESR genes are shown in Figure 1. For ESR1 gene, two blocks of 5 kb and 10 kb were identified with high LD. For ESR2 gene, only one 40 kb block was identified. For the case/control association tests, allelic and haplotypic analyses were conducted with chi-square test by Haploview. An empirical p<0.05, corrected after 5,000 permutations, was considered to be statistically significant. Once a significant association appeared for allele analysis, further genotypes analyses based on dominant and recessive models were conducted using logistics regression in IBM SPSS 21 with age, intellectual quotient (IQ), and gender as covariates.
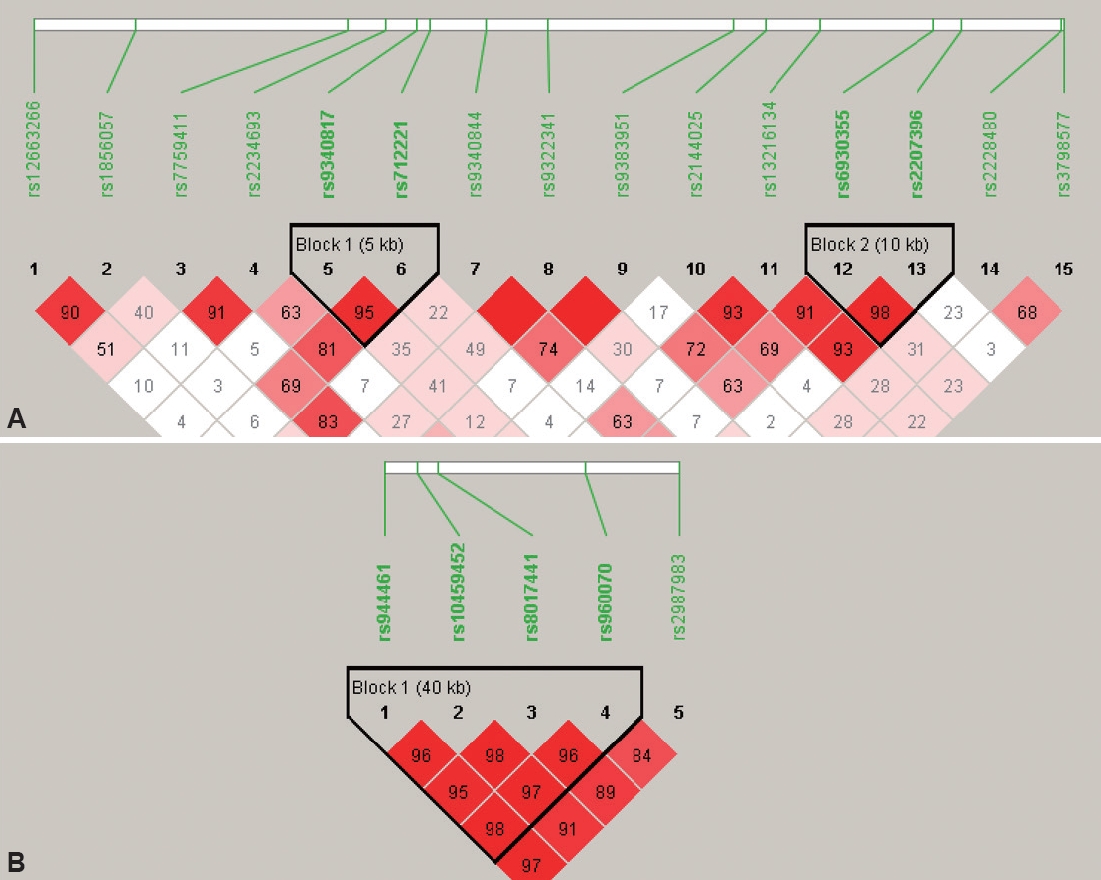
The linkage disequilibrium (LD) patterns of ESR1 (A) and ESR2 (B) genes. LD, measured as D’, was calculated using Haploview program (version 4.2; https://www.broadinstitute.org/haploview/downloads). Squares in red means strong LD. Numbers in bracket indicate the length of blocks.
Analysis of gene-gene interaction
Gene-gene interactions were examined by generalized multifactor dimensionality reduction (GMDR). A logistic regression model was used for the gene-gene interaction in ADHD presence, adjusted for age, gender, and IQ. The top-ranked interaction model for a given order interaction was chosen based on: 1) cross validation consistency (CVC) of ≥8/10, 2) prediction accuracy of >50%, and 3) empirical p<0.05 using 1,000 permutation tests. The samples were then divided into highrisk and low-risk combination groups in statistic, ignoring the plausible biological bases. After that, the significant interaction model from GMDR was verified by logistic regression and analyses of covariance using IBM SPSS 21.
RESULTS
Demographic data
A total of 1,997 participants were included: 1,035 in the ADHD group and 962 controls. The demographic characteristics of the subjects are shown in Table 2. The median age of the ADHD group was 109 (93–134) months, and 132 (105–264) months for the control group. A significantly smaller month age was observed in the ADHD group than the control group (p<0.001). Similarly, IQ was significantly lower in the ADHD group (103.91±14.74 vs. 112.73±14.08, p<0.001). There was a statistically significant difference in the gender distribution between the two groups: more males were in the ADHD group than the control group (84.3% vs. 63.1%, p<0.001).
Case/control study
Allelic and genotypic analyses
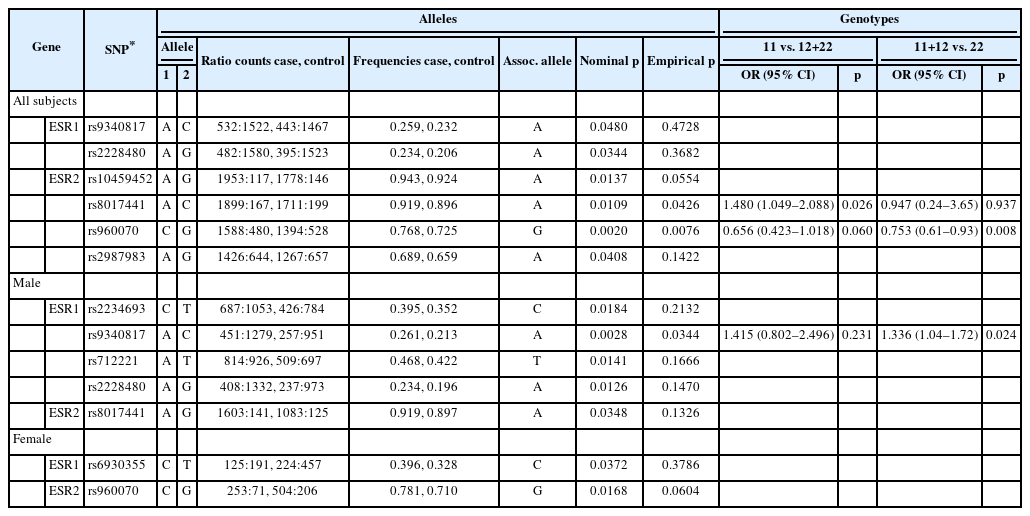
Table 3 summarized the association test results for SNPs with ADHD presence. For all subjects, no significant associations were found between two SNPs of ESR1 and ADHD. The A allele of rs9340817 and A allele of rs2228480 had nominal p< 0.05 but were not significant after 5,000 permutations (empirical p>0.05). For ESR2, the A alleles of rs8017441 (empirical p=0.0426) and rs960070 (empirical p=0.0076) were significantly associated with ADHD presence after permutation correction. The other two SNPs, rs10459452 (nominal p=0.0137, empirical p=0.0554) and rs2987983 (nominal p=0.0408, empirical p=0.1422) were significantly associated with the occurrence of ADHD prior to correction.
In addition, the association between SNPs of ESR genes and ADHD presence was also analyzed by gender (Table 3). In males, only ESR1 gene rs9340817 A allele was significantly associated with ADHD presence (empirical p=0.0344). The pvalue of ESR1 gene rs2234693 C allele and rs712221 T allele, ESR2 gene rs8017441 A allele were significant before permutations but did not remain statistically significant after correction. In females, no significant SNPs were found to be associated with ADHD after correction.
Genotypic analyses also support association between above SNPs with ADHD. We found that the AA genotype of rs8017441 (p=0.026, OR=1.480 [1.049–2.088]), GG genotype of rs960070 (p=0.008, OR=0.753 [0.61–0.93]) were more frequent in ADHD cases than in controls in all subjects. In males, CC genotype of rs9340817 (p=0.024, OR=1.336 [1.04–1.72]) was less frequent in ADHD cases, suggesting it may be a protective factor.
Haplotypic analyses
The results of haplotypic analyses in all subjects showed that the frequency of the TGGC haplotype (rs944461-rs10459452-rs8017441-rs960070) of ESR2 gene in ADHD cases was lower than that in controls (empirical p=0.0268) (Table 4). In male, the frequency of the AT haplotype (rs9340817-rs712221) of ESR1 in ADHD cases was higher than that in controls (empirical p=0.0058) (Table 4). No significant difference of haplotype between ADHD cases and controls was found in females after permutations.
Gene-gene interaction and the susceptibility of ADHD
All SNPs of the three neurodevelopmental genes and SNPs of ESR genes with nominal p<0.05 in case/control study were included for further gene-gene interaction analysis. When SNPs were in linkage disequilibrium, the SNP with the lowest p-value was included. The included SNPs were: rs2234693, rs9340817, rs6930355, and rs2228480 of ESR1; rs960070 and rs2987983 of ESR2; rs6265, rs2049046, and rs2030324 of BDNF; rs362987 of SANP25; rs6565113 of CDH13.
In the whole sample, no statistically significant interaction model was found between ESR genes and neurodevelopmental genes (p>0.05). In males, a five-locus SNP-SNP interaction model, which composed of ESR2/rs960070, BDNF/rs6265 and BDNF/rs2049046, SNAP25/rs362987, and CDH13/rs6565113, was significantly associated with the presence of ADHD (testing accuracy=54.57%, CVC=8/10, adjusted p<0.001). Details were shown in Table 5 and Figure 2. In females, a three-locus SNPSNP interaction model, which composed of ESR2/rs960070, BDNF/rs6265, and BDNF/rs2049046, was significantly associated with the presence of ADHD (testing accuracy=63.82%, CVC=8/10, adjusted p<0.001). Details were shown in Table 5 and Figure 3.
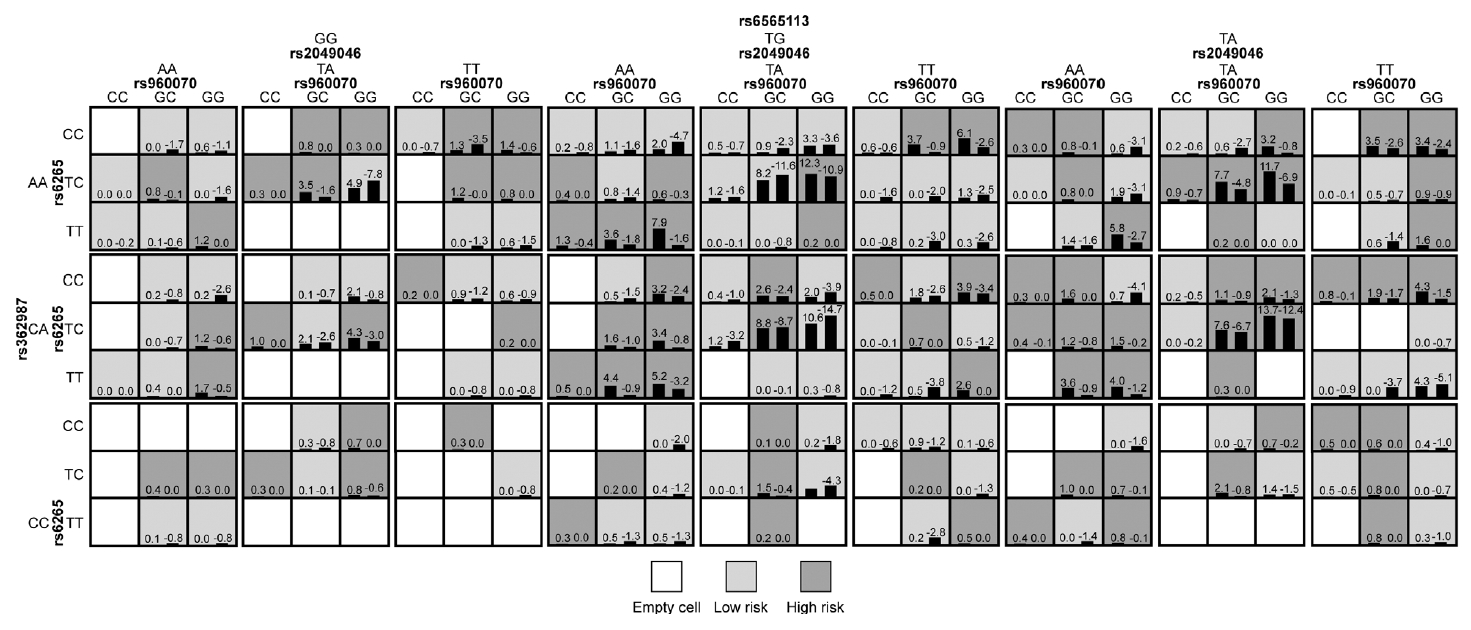
The identified best interaction model in male subjects showed a significant associated with ADHD presence (p<0.001) by GMDR. The left bar in each cell reflects a positive score, while the right bar represents a negative score. Dark shading indicates high-risk cells, light shading indicates low-risk cells, and no shading indicates empty cells. It is worth noting that the patterns of high-risk and low-risk cells alter across each of the distinct multi-locus dimensions, indicating epistasis. ADHD, attention deficit hyperactivity disorder; GMDR, generalized multifactor dimensionality reduction.
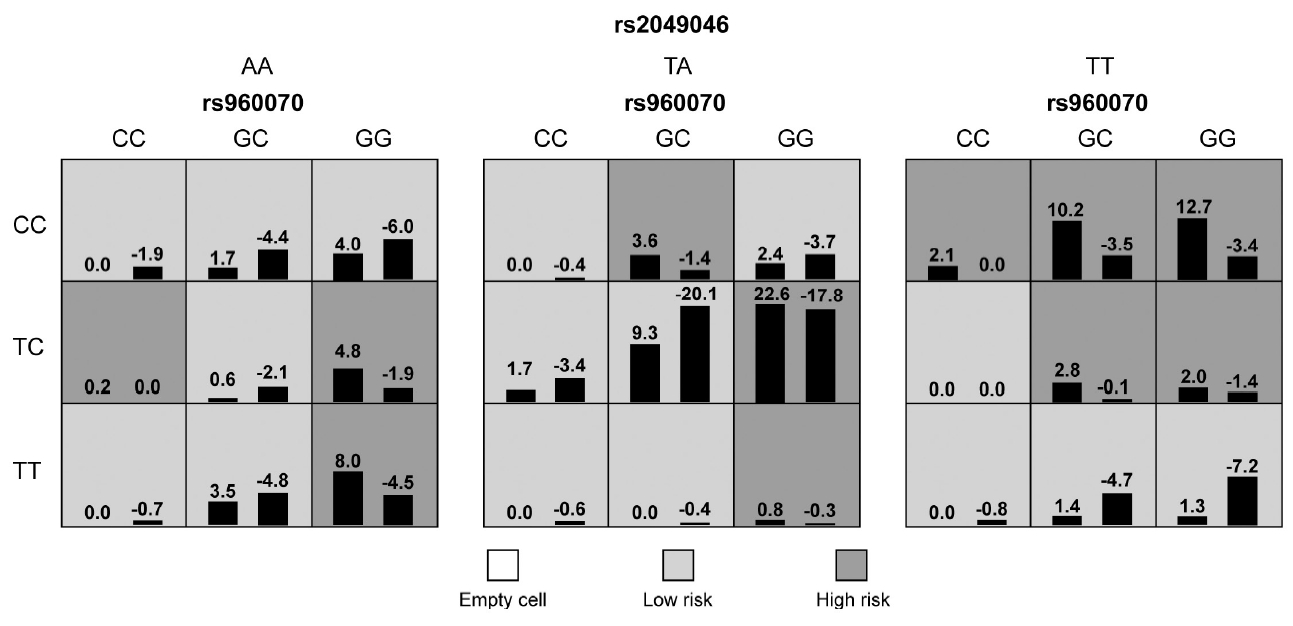
The identified best interaction model in female subjects showed a significant associated with ADHD presence (p<0.001) by GMDR. The left bar in each cell reflects a positive score, while the right bar represents a negative score. Dark shading indicates high-risk cells, light shading indicates low-risk cells, and no shading indicates empty cells. ADHD, attention deficit hyperactivity disorder; GMDR, generalized multifactor dimensionality reduction.
To verify the above models, subjects were then divided into low-risk and high-risk group according to the GMDR results. As shown in Figures 2 and 3, the light grey cells indicate genotype combinations of the low-risk group and dark grey cells indicate genotype combinations of the high-risk group. After adjusted for month, age, and IQ, the results of logistic regression analyses confirmed the significant difference between low-risk and high-risk group for both models (Table 6).
DISCUSSION
In this case/control study, we investigated the association between the polymorphism of ESR genes and the susceptibility of ADHD among Chinese Han descent. We identified several ESRSNPs related to ADHD. To our knowledge, this is the first study to discover ESRgene SNPs associated with ADHD presence. We also explored the gene-gene interaction effects on ADHD using six screened ESRgene SNPs and five selected neurodevelopmental gene SNPs. Valid interaction models were found for both males and females.
The most important finding of the present study lay in the significant association between polymorphism of ESRgenes and ADHD susceptibility. In all subjects, the polymorphism of ESR2 gene rs960070 and rs8017441 were found significantly related to ADHD presence. Moreover, ESR1 gene rs9340817 was observed associated with male ADHD. These finds were further validated in the case/control tests of genotype and haplotype. However, the impact of ESRgene polymorphism on ADHD is less well-studied presently. No study linked these three SNPs to ADHD. We also retrieved DisGeNET and GWAS Central for these three SNPs in ADHD and other mental disorders, but found no evidence [36,37]. Our data also suggested a trend of association between several SNPs and ADHD susceptibility: ESR1 rs2234693, rs712221, rs6930355, rs2228480, and ESR2 rs10459452, rs2987983. However, these SNPs were not found related to ADHD previously. While rs2234693 was not reported in ADHD, it has been linked to a variety of behavioral phenotypes and clinical outcomes, including anger expression in girl, children and adolescent anxiety, female depression, and etc [15,38-41]. A previous study indicated that rs2144025 is related to the comorbid psychological symptoms of ADHD in both sex. In contrast, the present study revealed no connection between rs2144025 and ADHD susceptibility in either gender [42]. In a study predicting ADHD severity from psychosocial stress and stress-response genes which utilized random forest, rs985191, rs77714417, rs74325817, rs35365822, rs9340910, and rs6930114 of ESR1 gene were ranked in the 25 highest-ranked predictors [43]. All these aforementioned SNPs warrant further investigation in ADHD.
Another significant finding of this research is that the impact of ESRgene polymorphism on ADHD differs across gender. The present study indicated that ESR1 gene rs9340817 A allele and rs9340817-rs712221 AT haplotype are risk factors for ADHD presence in males but not females. Similarly, there was a trend for ESR1 gene rs6930355 to be associated with ADHD in females but not males. These findings align with previous studies showing that ESRgenes affect the sex heterogeneity seen in psychiatric disorders [44-46]. Sexual dimorphism in ADHD has also been reported in our previous studies [47,48]. Prior work exploring the role of genes in the sexual dimorphism of ADHD has mainly focused on sex chromosomes and sex hormones. ADHD symptoms increase in the week before menstruation and resolve during pregnancy. Additionally, with the fluctuation of estrogen level, central stimulants have different therapeutic effects on female ADHD patients [49]. In males, ESR1 gene polymorphisms are associated with aggressive behavior [50]. In animal studies, after ESR1 gene knockout, both female and male mice showed a decrease in BDNF transcription and protein levels, but only female mice showed upregulation of dopamine D1 receptor transcription and protein levels, while male mice did not [51].
Overall, our results indicate that ESRgenes may play a role in the pathophysiology of ADHD, though the specific mechanism remains unclear. We also found that gene-gene interactions among ESR2 rs960070, BDNF rs6265 and rs2049046, SNAP25 rs362987, and CDH13 rs6565113 may be a risk factor for ADHD in Chinese Han males, and female ADHD was significantly associated with interaction model composed of ESR2 rs960070, BDNF rs6265 and rs2049046. These findings served as a reminder that neurodevelopmental and ESRgenes may jointly contribute to ADHD.
Multiple studies have indicated that estrogen can play a neuroprotective role through ESR[52]. Research has shown that estrogen signaling pathways protect the brain from inflammation and stress by promoting BDNF production [53]. Küppers et al. [51] observed decreased BDNF expression in the midbrain of ESR1 knockout mice. The role of BDNF rs6265 in the susceptibility of ADHD is controversial across studies. Luo et al. [54] reported that BDNF rs6265 does not seem to play a significant role in ADHD children, while Ozturk et al. [55] reached the opposite conclusion. Although BDNF rs2049046 was not reported previously in ADHD, He et al. [56] suggested that this SNPs may interact with job burnout. Both rs6265 and rs2049046 were related to the serum BDNF level, which can be influenced by the estrogen signaling pathways and exert its biological effect. For SNAP25, the gene has been found to be associated with susceptibility to ADHD [26]. Our previous study also found SNAP25 as a part of an interaction model contributed to susceptibility to ADHD in males [57]. The SNAP25 rs362987 polymorphism has been linked to ADHD, possibly by affecting the exocytosis of the neurotransmitter gamma-aminobutyric acid (GABA) [58]. Estrogen can influence brain morphology by changing GABA (A) receptor expression and function, which future influence learning and memory [59,60]. For CDH13, rs6565113 polymorphism tended to be associated with specific phenotypes of ADHD [34]. The interaction of ESRgenes and the above neurodevelopmental genes may be a possible mechanism contributing to the effect of ESRgene on ADHD susceptibility.
This study still has the following limitations. First, the number of SNP locus from ESRgenes included in this study was relatively small, which may not fully reflect the role of their polymorphisms in ADHD, and important areas may be missed. Second, it is unclear whether the sample size and power sufficient for identifying gene-gene interactions, so we verified results of this part by logistic regression analysis. Certainly, expanding the sample size cloud make the results more reliable. Furthermore, the serum levels of ESRand other biomarkers were not assessed in this study, which could have provided additional insights. In future studies, measuring serum level of biomarkers and including a larger number of ESRgene SNPs could help validate and expand upon these initial findings.
In conclusion, the present study investigated the ESRgene polymorphisms in Chinese Han children with ADHD, and found they are closely associated with susceptibility of ADHD. Furthermore, we also reveal that the genetic interaction among ESRgenes and the neurodevelopmental genes (i.e., BDNF, SNAP25, and CDH13) plays an important role in ADHD. These findings can help explain the gender difference of susceptibility of ADHD, but still need future validation.
Notes
Availability of Data and Material
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors have no potential conflicts of interest to disclose.
Author Contributions
Conceptualization: Yiwei Lin, Haimei Li, Lu Liu, Qiujin Qian. Data curation: Yiwei Lin, Haimei Li, Lu Liu. Formal analysis: Yiwei Lin, Haimei Li, Qiujin Qian. Funding acquisition: Haimei Li, Lu Liu, Qiujin Qian. Methodology: Yiwei Lin, Lu Liu. Writing—original draft: Yiwei Lin, Qiujin Qian. Writing—review and editing: all authors.
Funding Statement
This study was partly funded by the National Basic Research Development Program of China (grant number 973 program 2014CB846104), National Natural Sciences Foundation of China (grant numbers 81071109; 81301171).
Acknowledgements
We would like to express our sincere gratitude to Dr. Hao Zhao for his invaluable assistance in data analysis and expert utilization of the program.