Disrupted-in-Schizophrenia-1 SNPs and Susceptibility to Schizophrenia: Evidence from Malaysia
Article information
Abstract
Objective
Even though the role of the DICS1 gene as a risk factor for schizophrenia is still unclear, there is substantial evidence from functional and cell biology studies that supports the connection of the gene with schizophrenia. The studies associating the DISC1 gene with schizophrenia in Asian populations are limited to East-Asian populations. Our study examined several DISC1 markers of schizophrenia that were identified in the Caucasian and East-Asian populations in Malaysia and assessed the role of rs2509382, which is located at 11q14.3, the mutual translocation region of the famous DISC1 translocation [t (1; 11) (p42.1; q14.3)].
Methods
We genotyped eleven single-neucleotide polymorphism (SNPs) within or related to DISC1 (rs821597, rs821616, rs4658971, rs1538979, rs843979, rs2812385, rs1407599, rs4658890, and rs2509382) using the PCR-RFLP methods.
Results
In all, there were 575 participants (225 schizophrenic patients and 350 healthy controls) of either Malay or Chinese ethnicity. The case-control analyses found two SNPs that were associated with schizophrenia [rs4658971 (p=0.030; OR=1.43 (1.35-1.99) and rs1538979-(p=0.036; OR=1.35 (1.02-1.80)] and rs2509382-susceptibility among the males schizophrenics [p=0.0082; OR=2.16 (1.22-3.81)]. This is similar to the meta-analysis findings for the Caucasian populations.
Conclusion
The study supports the notion that the DISC1 gene is a marker of schizophrenia susceptibility and that rs2509382 in the mutual DISC1 translocation region is a susceptibility marker for schizophrenia among males in Malaysia. However, the finding of the study is limited due to possible genetic stratification and the small sample size.
INTRODUCTION
Schizophrenia is a complex set of disturbances in thinking, perception, affect, and social behaviour. The causes of this disorder are still largely unknown. Significant family, twin, and adoption studies have indicated that genetic factors play an important role in the etiology of schizophrenia. Since the discovery of the Disrupted-in-Schizophrenia-1 (DISC1) gene within a balance translocation (1; 11) (q42.1; q14.3) in a large Scottish family with psychiatric disorders1 more than 20 years ago, there have been many studies, especially in Caucasian populations, which have attempted to identify specific DISC1 variants association with Schizophrenia.2,3 The evidence of the association of the DISC1 gene with schizophrenia in several Caucasian populations, especially in Europe,4,5 as well as the Caucasian populations that reside outside the European continent,6 is positively encouraging. However, the recent data from genome-wide scans mega meta-analysis studies have cast doubt on this association.7
In contrast to the inconclusive evidence for DISC1 as a genetic risk factor for schizophrenia in Caucasian populations, functional and cell biology studies have consistently supported the connection between the DISC1 gene and neuropathological theories of schizophrenia. DISC1gene is involved in a variety of neurocellular functions, such as intracellular signaling, neurodevelopment, and neurosynapses.8,9,10,11,12 Functionally, DISC1gene mutations and various haplotypes are associated with impaired memory.13,14 Rats with DISC1 gene knockout were found to exhibit stress-induced prefrontal cortex dysfunction,15 an important precondition for the development of schizophrenia.
Because genetic variations differ among populations, it is important to assess the variations of DISC1 gene role in schizophrenia in a local context before making a definitive decision regarding its role in schizophrenia. Regrettably, in Asia, there had been only few studies that have investigated the role of the DISC1 gene in Schizophrenia, and these studies were mainly performed on East-Asian populations.16,17,18,19,20 There were conflicting findings on the association between schizophrenia and DISC1 in Chinese16,17 and Korean populations18,19 whilst a study on DISC1/TRAX locus found insignificant linked of the locus in the Japanese.20 In addition to the DISC1 locus, the mutual translocated regions of the DISC1 gene (1; 11) (q42.1; q14.3) are also areas of interest. A recent meta-analysis of several genome-wide studies that also included publicly available data in the Genetic Association Information Network (GAIN) (www.genome.org) identified rs2509382 as being associated with schizophrenia among males.21 This is an interesting finding because rs2509382 is located at 11q14.3 and represents the mutual translocation region.
In the present study, a case-control association analysis was performed to examine several DISC1 markers of schizophrenia that were identified in Caucasian and East-Asian populations in terms of their relevance to Malaysian patients. The study also assessed the role of rs2509382 among the patients.
METHODS
Ethics approval
The study was registered under the National Medical Research Registry of Malaysia (NMRR-10-832-6366), and the protocol was approved by both the Medical Research and Ethics Committee, Ministry of Health, Malaysia, and the Kulliyyah of Medicine, International Islamic University, Malaysia (IIUM).
Study populations
The study consisted of 225 unrelated schizophrenia patients and 350 healthy controls. Schizophrenia cases were collected from the Psychiatry Clinic of Tengku Ampuan Afzan Hospital, Kuantan (HTAA). The patients who attended follow-up appointments at the clinic were either from Kuantan (the state capital of Pahang) or referred from other districts in Pahang or the town Kemaman (in Terengganu). The control subjects were from the district of Kuantan. All subjects were over 18 years of age and were legitimately identified as being of Malay or Chinese ethnicity based on their personal identification card issued by the National Registry Department of Malaysia. These subjects also had grandfathers and grandmothers of similar ethnicities. The schizophrenic subjects had a consensus best-estimate DSM-IV diagnosis of schizophrenia of at least six months duration, and the symptoms of psychosis were not deemed to be secondary to substance use or neurological disorders. Mentally retarded patients were not included as subjects. In all, there were 225 (147 males and 78 females) schizophrenic patients and 350 controls (233 males and 117 females), with those of Chinese ethnicity representing only 43 and 10 of the cases and control subjects, respectively. The sample size was calculated by using OpenEpi for the unmatched case-control study.22 Using data from Schumacher et al.2 for rs4658890 (MAF=0.485), a total sample size of 994 (1:1 case-control ratio) was needed for a standard 95% two-sided confidence level and an 80% power of detection (effect size 0.30). In our study, a total of 225 schizophrenia patients and 350 controls would give less than a 60% power of detection at a 95% confidence level.
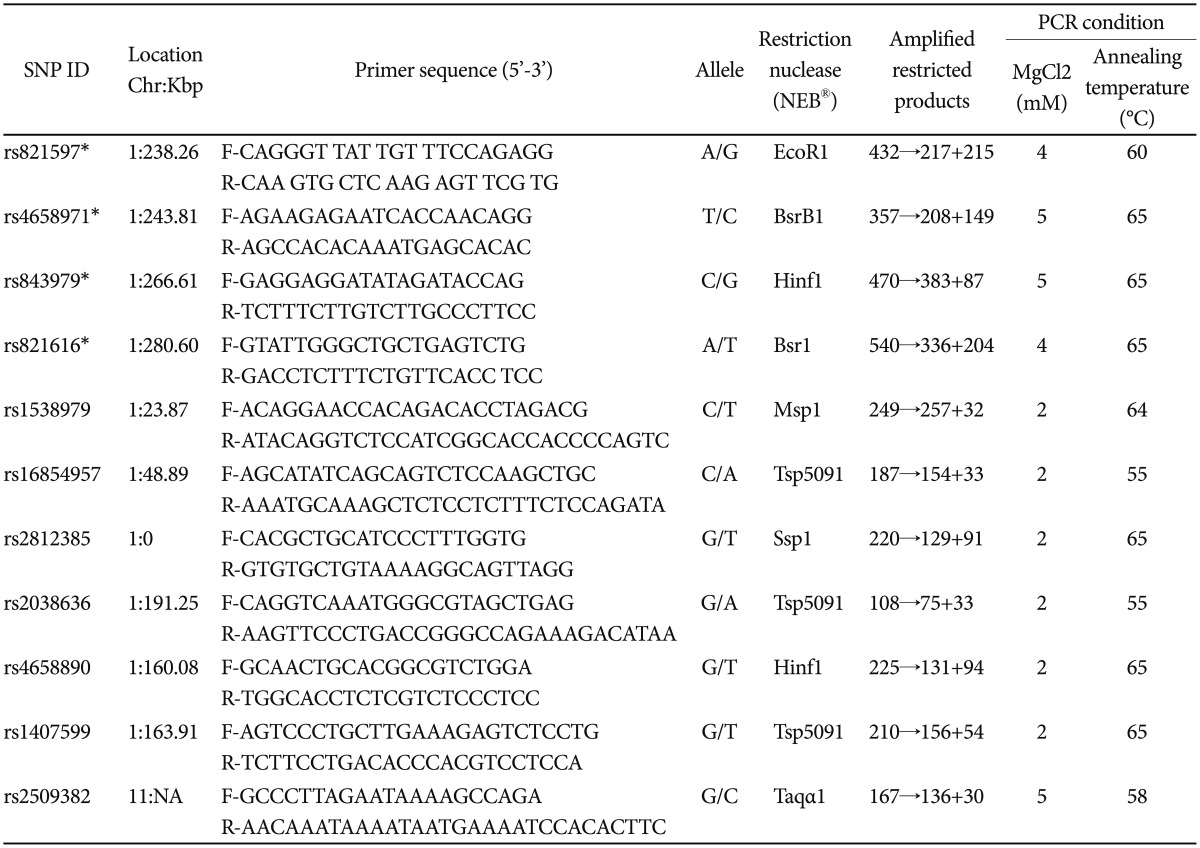
Genotyping and selection of SNPs
Peripheral blood samples were collected from all 575 subjects. Genomic DNA was extracted using a Gentra® Puregene® (Qiagen, Germany) purification kit, following the manufacturer's protocol. Four SNPs (rs821597, rs4658971, rs843979, and rs821616) that were located in the region between Intron 10 and Exon 11 of DISC1 were selected based on a previous study that was conducted on Han Chinese subjects.17 Another six SNPs (rs1538979, rs2812385, rs4658890, rs1407599, rs16854957, and rs2038636) were selected based on the meta-analysis findings of DISC1 studies performed among Caucasians in Europe.2 The last SNP, rs2509382, which was located at 11q14.3, represented the mutual translocation region of the famously translocated DISC1 (1q42.1) gene.21 The intermarker linkage disequilibrium patterns of the SNPs are shown in Figure 1.
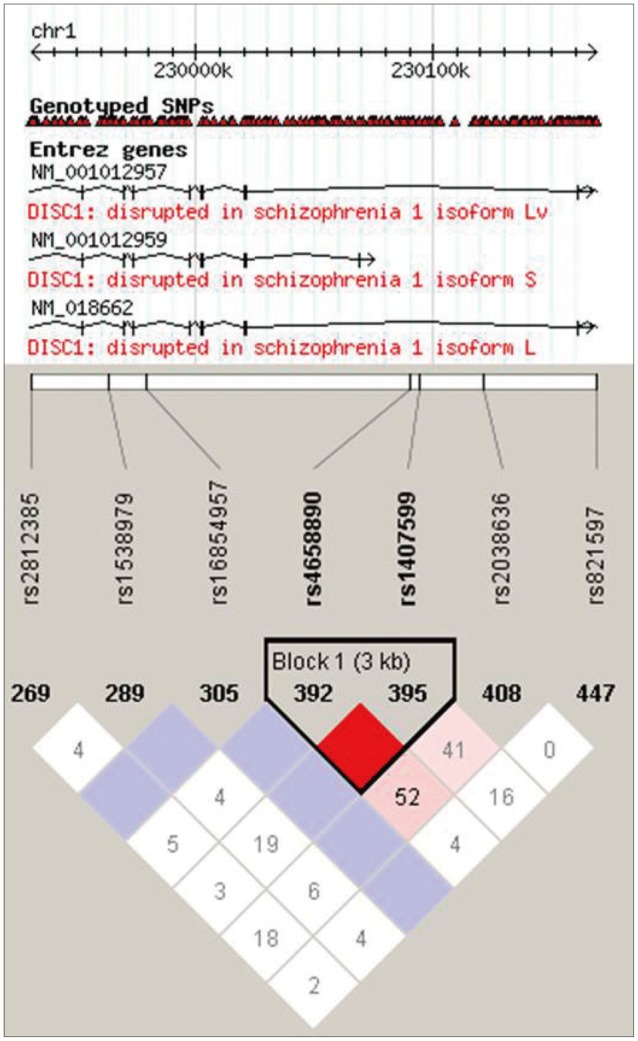
Lingkage disequilibrium between the SNPs selected in the DISC1 gene constructed using Haploview 4.2 of HCB Chr 1: 229329..230397k database. The rs4658971, rs843979 and rs821616 were uncaptured. Values are equal to D' multiplied by 100 (Values are not shown for D'=1). SNP: single nucleotid polymorphism.
SNPs were genotyped using PCR-RFLP methods. PCR was performed in a 15 µL reaction volume that included 0.25 µM of each primer, 200 µM of dNTPs, I U of One Taq™ Polymerase (New England Biolabs), 1X reaction buffer of various MgCl2 titrations, and 30 ng of DNA. The information regarding the primers and restriction enzymes used in the assay is shown in Table 1. The PCR products were digested with their respective restriction enzymes, followed by electrophoretic separation on 3.2% agarose gel. Only clear and unequivocal genotypes were included in the data analysis. Thus, the PCR-RFLP success rate of most of the SNPs studied ranged between 85% and 99%. Only one SNP (rs843979) had success rate of 73%. A minimum of 3 samples per SNP, each representing the ancestor, variant and mixed genotypes were randomly chosen based on the RFLP result, for direct nucleotide sequencing. The concordance rate between RFLP and sequencing was 100%.
Statistical analyses
The analyses of the allelic and genotypic differences in DISC1 gene markers between schizophrenic and control subjects were evaluated via Chi-square test, which was performed using the Shesis software program.23 The odds ratio (OR) for the alleles' effect and the deviation of the genotype counts from the Hardy-Weinberg equilibrium were tested using the same software. A Bonferroni correction for multiple testing was carried out to exclude type I errors (corrected significant p of <0.005). Data from studies performed among the Chinese16,17 and Japanese populations24 were used for meta-analyses. The meta-analyses and heterogeneity tests were performed using the Comprehensive Meta-analysis Version 2 software program.25
RESULTS
Ten SNPs within the DISC1 gene and rs2509382 in the DISC1 mutual translocated region of 11q14.3 were genotyped in 225 schizophrenic patients and 350 healthy controls from Malay-Malaysian and Chinese-Malaysian populations. Genotype testing on two SNPs (rs2038636 and r16854957) found they were not polymorphic in the populations studied. None of the controls and cases showed heterozygous or homozygous variants. The genotypic and allelic differences between schizophrenic patients and controls for the remaining nine SNPs are shown in Table 2. The distributions of all of the SNP genotypes conformed to Hardy-Weinberg equilibrium in the control samples.
There were significant differences in genotypes and allele frequencies between patients and controls for two SNPs, rs4658971 and rs1538971. Those having the T minor allele instead of the C major allele for rs4658971 [p=0.0303, OR=1.44 (1.03-2.00)] and rs1538971 [p=0.036, OR=1.35 (1.02-1.80)] are more susceptible to schizophrenia. Except for significant genotypic differences for rs4658971 (χ2=14.66, p=0.00067), the genotypic and allelic differences were reversed after a Bonferonni correction for multiple testing. The nine SNPs were further tested for a relationship with disease based on the gender. The results of the genotype and allele frequencies for males and females are shown in Table 3 and 4, respectively. There were significant differences in genotype (p=0.0304) and allele frequencies in male participants for SNP rs2509382 [p=0.0082, OR=2.16 (01.22-3.81); corrected p=0.090].

Association of genotypes and alleles distribution of nine DISC1 related SNPs with schizophrenia in males

Association of genotypes and alleles distribution of nine DISC1 related SNPs with schizophrenia in females
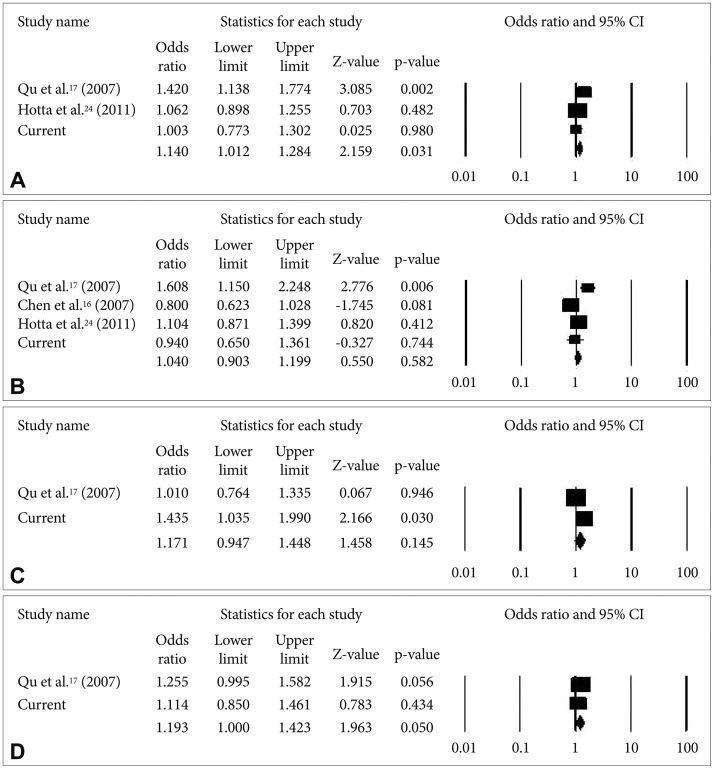
The meta-analyses results for rs821616, rs821597, rs843979, and rs4658971 are shown in Figure 2. There was a significant association between rs821597 and schizophrenia in the combined data [p=0.031, OR=1.14 (1.01-1.28); heterogeneity test (I2=62.6, p=0.069)]. In contrast, the meta-analysis of rs4658971 found no significant association of this SNP with schizophrenia.
DISCUSSION
There are conflicting findings regarding the association of the DISC1 gene with schizophrenia. This inconsistency could be due to the limited selection of polymorphic markers that were tested or the inadequacy of the sample's representation. Based on the summary of all published association studies of the DISC1 gene and schizophrenia that were accessed in the Schizophrenia Research Forum (www.szgene.org),26 there were more than 25 studies performed on Caucasian populations, as compared to less than eight performed on Asian populations.
In Asia, there have been studies that showed an encouraging association between the DISC1 gene and schizophrenia among the Han Chinese.16,17 A study among the Japanese20 and Korean populations,18 however, showed that DISC1 gene variants had a negative role in schizophrenia. Nevertheless, a study of the Korean population also indicated that DISC1 variation is associated with the poor-concentration phenotype of schizophrenia.19 The two SNPs (rs821616 and rs821597) that were reportedly associated with schizophrenia17 showed no such association in our cohort of patients. However, this study found two SNPs (rs4658971 and rs1538979) to be significantly associated with schizophrenia in terms of both genotypes and allelic distribution. The effect of these variations in DISC1 gene expression are unknown because both variations are located within introns of the DISC1 gene. There is no evidence suggesting the effects of these two SNP on gene splicing. Interestingly, a meta-analysis of more than five case-control association studies performed in Europe found that rs1538979 was a susceptibility marker for schizophrenia.2 However, an upgrade meta-analysis studies3 has refuted the association of the common variants of DISC1 with schizophrenia.
A recent meta-analysis of several genome-wide studies, including the publicly available data of the Genetic Association Information Network (GAIN) (www.genome.org), identified rs2509382 as having an association with schizophrenia among males.21 Our study found that rs2509382 caused a similarly significant risk of schizophrenia among the males [p=0.0082, OR=2.16 (01.22-3.81)]. This finding supported the possibility of a sex-specific genetic predisposition toward the development of schizophrenia. Interestingly, rs2509382 is located at 11q14.3 near the non-coding and brain expressed DISC1Fusion Partner 1 (DISC1FP1) gene27 that represents the partner of translocation region of the famously described DISC1 (1q42.1) translocation. The DISCIFP1 region association with schizophrenia is not well explored although a third party analysis on the Psychiatric Genetics Consortium Genome-Wide Association Study (GWAS) data has reported a weak association.28 The exploration of the association of other variants within the 11q14.3 location with schizophrenia may provide further information regarding the relationship between this genetic location and schizophrenia.
Our study analyzed both Malay and Chinese subjects as one cohort of participants. This was based on the assumption that the genetic variations among the Malay and Chinese populations were relatively similar, as suggested by the Singapore Genome Variation Project,29 which was performed in a neighbouring country, Singapore. Additionally, evidence from a study on the genetic structure of populations found that all sub-ethnic groups of the Peninsular Malay are clustered together with the Chinese population and that these clusters are quite separated from the Yoruba, Indians, and Malaysian Indigenious Negritos.30
A meta-analysis was performed to improve the statistical power of this project by combining data from several studies performed on the Chinese population. Combining allele frequency results from three studies, we found that there was no significant evidence of an association between SNPs rs821616, rs843979, and rs4658971 and schizophrenia. However, the combined data showed that rs821597 carries the risk of susceptibility to schizophrenia. The current genetic evidence further supports the important role of the DISC1 gene in the development of schizophrenia, and the association seems to affect both Caucasian and Asian populations.
Several functional and cell biology studies have supported the role of DISC1 in schizophrenia. DISC1 product interacts with other proteins implicated in the central nervous system development.10,31 In-vitro, DISC1 has been shown to have a multifunctional protein whose truncation may lead to the disruption of intracellular transport, neurite growth, and neuronal migration.28 In addition to neurodevelopment, other studies also revealed the functional roles of DISC1 in transcription and cAMP signalling32 and in the presynaptic activity of glutamate release.33 The positive findings regarding genetic predisposition found in the current study, combined with the implicated proteomic pathways and functional roles of DISC1 that were reported by various studies, give additional support to the importance of DISC1 in the pathology of schizophrenia.
Our study has several limitations. The case-control approach used in this study is prone to false positive and false negative associations, especially due to undeclared population stratifications. The Malay ethnic group is loosely defined based on Article 160 of the Constitution of Malaysia. Therefore, a Malay is defined as a Malaysian citizen born to a Malaysian citizen who professes to be a Muslim, habitually speaks the Malay language, and adheres to Malay customs. To reduce this bias, the Malay participants were identified based on the ethnic identification of the previous two generations. Because schizophrenia is a disease with complex clinical presentations, a general association based on either having or not having the disease, as was used by the current study, may be too simplistic to unravel the complex nature of the disease with regard to genetic predisposition.
In conclusion, the current study found two SNPs within the DISC1 gene that predispose individuals to schizophrenia within the Malaysian population. Additionally, we found an association of rs2509382 with schizophrenia among male participants, thus reinforcing previous genome-wide meta-analysis findings regarding this SNP in a male Caucasian population.21 Additional studies that investigate the interaction between genetic predisposition and the clinical and social features of schizophrenia could elucidate the role of this gene in the clinical pathology of schizophrenia.
Acknowledgments
Staff and patients of Psychiatry Clinic, Tengku Ampuan Afzan Hospital, Kuantan, Pahang & staff of Molecular Research Laboratory, Kulliyyah of Medicine, IIUM. This study was supported by ERGS Grant (ERGS-13-027-0060) by the Ministry of Education, Malaysia. The authors have no conflicts of interest that are directly relevant to the content of this manuscript.
The study is funded by the ERGS Grant (13-027-0060) provided by the Ministry of Education, Malaysia.